Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by SAMSON ANOSH BABU P.
Early detection and analysis of lung cancer involve a precise and efficient lung nodule segmentation in computed tomography (CT) images.
- segmentation
- deep learning
- computed tomography
1. Introduction
According to data released by the World Health Organization (WHO) on 3 February 2020, cancer is one of the leading causes of premature death in 134 of 183 countries. It has been observed that, especially in 2018, most of the prominent cancer deaths are due to lung cancer (1.76 million deaths). Detection and analysis of the lung nodules at an early stage facilitate efficient treatment and drastically improve a patient’s chance of survival [1]. CT scans are a widely used and highly accurate format for the purpose of screening and analyzing lung nodules, especially in differentiating the nodules from other structures. Moreover, the precise segmentation of these nodules is critical, considering the heterogeneity of the size, texture, location, and shape of the nodules, and the fact that their intensity may differ within the borders [2]. There are various types of lung nodules as observed in Figure 1 such as adhesion-type (juxtapleural and juxta-vascular), isolated, cavitary, calcified, small, and ground-glass opacity (GGO) nodules [3].
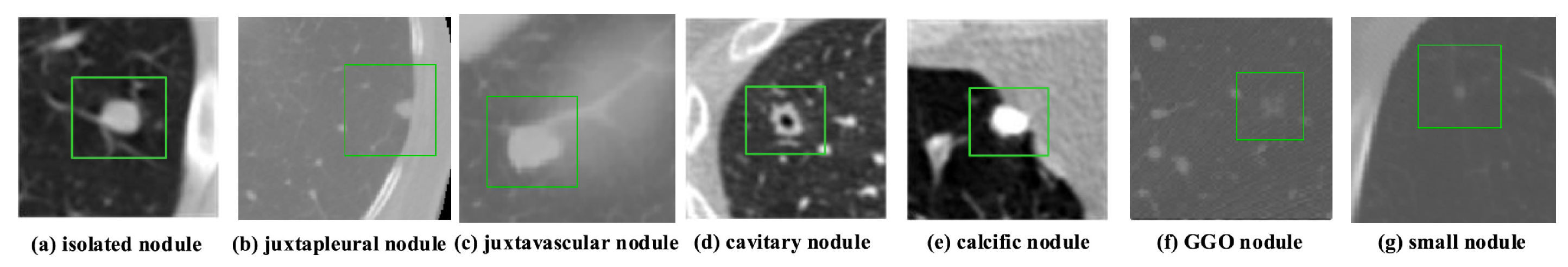
Figure 1. Illustrations of various types of lung nodules present in CT scans. Note: Small nodule indicates a nodule of diameter <6 mm. (a) isolated nodule; (b) juxtapleural nodule; (c) juxtavascular nodule; (d) cavitary nodule; (e) calcific nodule; (f) GGO nodule; (g) small nodule.
Another challenge lies in the segmentation of lung nodules, which is found in the case of nodules with small diameter and intensity comparable to that of the surrounding noise, which thereby hinders the down-sampling potential of the segmentation network, where the network cannot extract more in-depth semantic network features [4]. It significantly impacts the accuracy of the extraction of feature maps of large nodules. Based on these reasons, a robust segmentation network is necessary to accommodate the large-scale nodule (various types) problem.
Recently, convolutional neural networks (CNN) have become the mainstream architecture in the field of computer vision. One such architecture, the U-Net, is an encoder–decoder-like CNN architecture, which has shown exceptional results in segmentation of biomedical images [5]. Many modified U-Net architectures have achieved significant results in different domains of biomedical imaging. However, CNN architectures that are implemented for the task of lung nodule segmentation are still immature. Therefore, the development of advanced architectures dealing with the shortcomings of previous architectures is essential.
To deal with the challenges of efficient feature extraction and adaptation to heterogeneity of lung nodules, this preseaperrch proposes a modified U-Net architecture with a weighted bidirectional feature network (U-Det), which is appropriate for the segmentation of many forms of lung nodules. Figure 2 illustrates the pipeline of the proposed model.
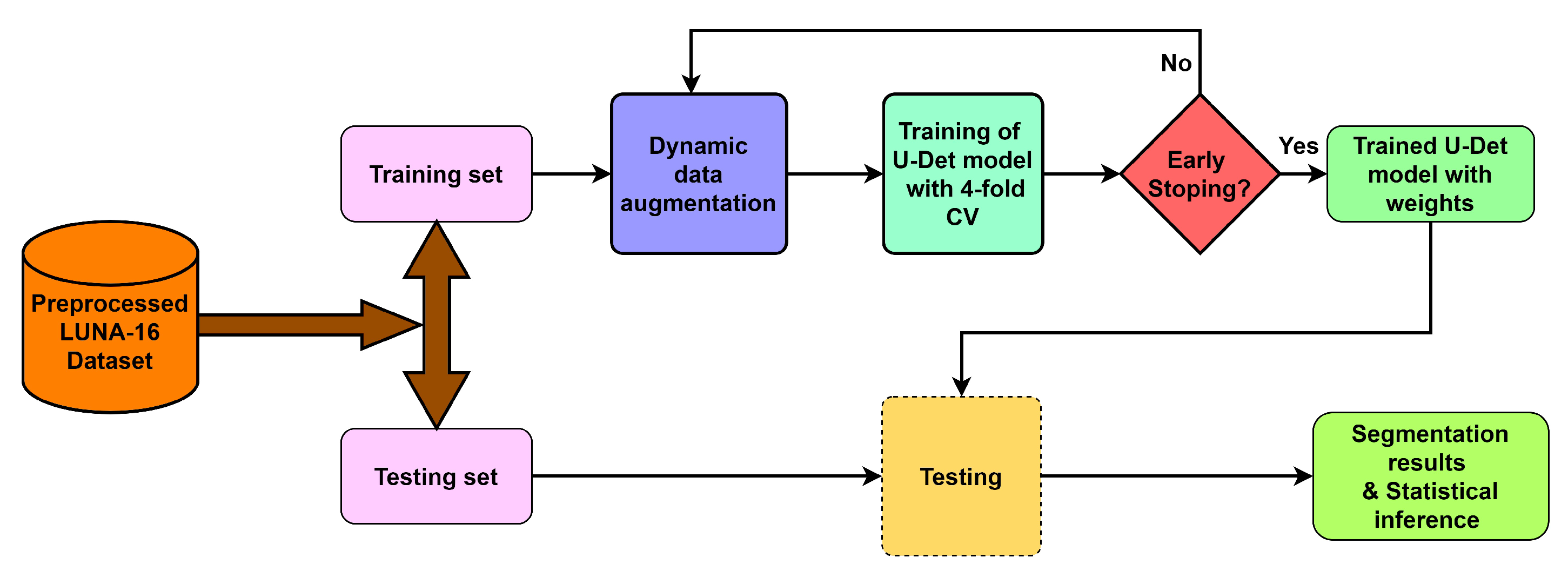
Figure 2. Overview of the proposed model pipeline.
2. Conventional Approaches
Many conventional approaches, such as morphological methods, region-growing processes, energy-based optimization techniques, and machine learning methods, were proposed for lung nodule segmentation in the literature. In morphological methods, morphology-based operations and those based on shape hypothesis were applied to isolate lung nodules by selecting the connected region [6]. However, the isolation of lung nodules using morphological operations did not perform efficiently [7]. Following that, region-growing methods were proposed to improve lung nodule segmentation but it was observed that these methods were unable to segment juxta-vascular and juxtapleural nodules and were only well suited to isolate calcified nodules [3]. In this case, for region-growing methods, the convergence condition and irregular-shaped nodules create difficulty due to the breach of the shape hypothesis. Similarly, energy based-optimization methods were also proposed for lung nodule segmentation. These methods consist of a level set function for the characterization of the image, and an energy function to typically turn the segmentation task into an energy minimization problem [8]. However, juxtapleural nodules and low contrast nodules such as GGO nodules drastically affect the improvement of lung nodule segmentation.
3. Machine-Learning-Based Approaches
In the past decade, many machine-learning–based methods were proposed for lung nodule segmentation [9]. An example of one such approach is the hybrid model for classifying the lung nodules with high-level feature maps proposed by Lu et al. [10]. Further, methods based on modified support vector machines were suggested to detect small lung nodules in 3D CT scans [11,12][11][12]. Down the line, researchers also developed 3D large-scale nodule segmentation techniques based on the Hessian strategy in combination with neural networks [13]. Similarly, segmentation approaches based on multi-phase models [14], unsupervised k-means clustering, and level set algorithms [15] were also proposed.
Among the recently developed deep learning-based approaches for segmentation, CNN is a multi-layered neural network that learns to map original image files and corresponding labels hierarchically, and to change the segmentation task into the classification of voxels [16,17][16][17]. For instance, Wang et al. introduced a multi-view CNN (MV-CNN) for nodule segmentation [18]. Further, Huang et al. developed a fast and fully automated detection and segmentation approach for pulmonary nodules in thoracic CT scans using deep convolutional neural networks [19]. Moreover, the authors Subham and Raman developed a lung nodule segmentation approach Using 3-dimensional convolutional neural networks [20]. A good review is also available on pulmonary nodule detection and diagnosis using deep learning applications in computed tomography images [21]. Moreover, Havaei et al. in [22] proposed a cascaded variant of CNN (Cascaded-CNN) for brain tumor segmentation. Further extending the use of CNN to lung nodule segmentation, Shen et al. proposed a multi-crop CNN (MC-CNN) to extract salient nodule features and classify malignancy [16]. Furthermore, Wang et al. in [18] introduced a multi-view deep CNN (MV-DCNN), and Sun et al. in [23] introduced a three multi-channel region of interest (RoI)-based CNN (MCROI-CNN) for lung nodule segmentation and displayed the effectiveness of CNN over existing computer-aided diagnosis systems. Further expanding work on CNN-based architectures, Zhao et al. have advocated an enhanced pyramid deconvolution network for increasing performance on lung nodule segmentation which blends low-level fine-grained characteristics with high-level functional characteristics [24].
On the other hand, fully convolutional networks [25] are a different approach for CT image segmentation. For example, the architectures 2D U-Net and 3D U-Net, proposed by Ronnerberger et al. and Ciccek et al., respectively, are better adapted to biomedical imaging [5,26][5][26]. The fully convolutional network U-Net (FCN-UNET) architecture is a convolutional network architecture used for fast and precise segmentation of images. Similarly, the central focused CNN (CF-CNN) proposed by Wang et al. is a data-driven method without involving the shape hypothesis. It showed strong performance for the segmentation of juxtapleural nodules [27]. Moreover, Cao et al. recently proposed incorporating intensity features into the CNN architecture by implementing a dual-branch residual network (DB-ResNet) [28].
Segmentation 2D convolutional U-Network (SegU-Net), a U-Net-based architecture, has also been suggested for lung nodule segmentation [29]. For pre-processing, Wang et al. and Cao et al. implemented weighted sampling of training data to deal with the comparatively smaller size of the Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IDRI) dataset [27,28][27][28]. The input slice of a CT scan is cropped to a smaller size with a random weighted sampling strategy to increase the size of the training dataset in the abovementioned method. Recently, Singadkar et al. proposed another approach toward lung nodule segmentation based on a deep deconvolutional residual network (DDRN) [30]. In DDRN, the approach was based on an encoder–decoder architecture with one-directional long-skip connections. Moreover, Wen-Fan Chen et al. developed a residual-dense-attention (RDA) U-Net network architecture for hepatocellular carcinoma segmentation [31].
References
- WHO. WHO Report on Cancer: Setting Priorities, Investing Wisely and Providing Care for All; WHO: Geneva, Switzerland, 2020.
- MacMahon, H.; Austin, J.H.; Gamsu, G.; Herold, C.J.; Jett, J.R.; Naidich, D.P.; Patz Jr, E.F.; Swensen, S.J. Guidelines for management of small pulmonary nodules detected on CT scans: A statement from the Fleischner Society. Radiology 2005, 237, 395–400.
- Kubota, T.; Jerebko, A.K.; Dewan, M.; Salganicoff, M.; Krishnan, A. Segmentation of pulmonary nodules of various densities with morphological approaches and convexity models. Med. Image Anal. 2011, 15, 133–154.
- Zhang, S.; Chen, X.; Zhu, Z.; Feng, B.; Chen, Y.; Long, W. Segmentation of small ground glass opacity pulmonary nodules based on Markov random field energy and Bayesian probability difference. Biomed. Eng. Online 2020, 19, 1–20.
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berling, Germany, 2015; pp. 234–241.
- Kuhnigk, J.M.; Dicken, V.; Bornemann, L.; Bakai, A.; Wormanns, D.; Krass, S.; Peitgen, H.O. Morphological segmentation and partial volume analysis for volumetry of solid pulmonary lesions in thoracic CT scans. IEEE Trans. Med. Imaging 2006, 25, 417–434.
- Diciotti, S.; Lombardo, S.; Falchini, M.; Picozzi, G.; Mascalchi, M. Automated segmentation refinement of small lung nodules in CT scans by local shape analysis. IEEE Trans. Biomed. Eng. 2011, 58, 3418–3428.
- Rebouças Filho, P.P.; da Silva Barros, A.C.; Almeida, J.S.; Rodrigues, J.; de Albuquerque, V.H.C. A new effective and powerful medical image segmentation algorithm based on optimum path snakes. Appl. Soft Comput. 2019, 76, 649–670.
- Dharmalingham, V.; Kumar, D. A model based segmentation approach for lung segmentation from chest computer tomography images. Multimed. Tools Appl. 2019, 79, 1–26.
- Lu, L.; Devarakota, P.; Vikal, S.; Wu, D.; Zheng, Y.; Wolf, M. Computer aided diagnosis using multilevel image features on large-scale evaluation. In Proceedings of the International MICCAI Workshop on Medical Computer Vision, Nagoya, Japan, 26 September 2013; Springer: Berling, Germany, 2013; pp. 161–174.
- Santos, A.M.; de Carvalho Filho, A.O.; Silva, A.C.; de Paiva, A.C.; Nunes, R.A.; Gattass, M. Automatic detection of small lung nodules in 3D CT data using Gaussian mixture models, Tsallis entropy and SVM. Eng. Appl. Artif. Intell. 2014, 36, 27–39.
- Prabukumar, M.; Agilandeeswari, L.; Ganesan, K. An intelligent lung cancer diagnosis system using cuckoo search optimization and support vector machine classifier. J. Ambient. Intell. Humaniz. Comput. 2019, 10, 267–293.
- Gonçalves, L.; Novo, J.; Campilho, A. Hessian based approaches for 3D lung nodule segmentation. Expert Syst. Appl. 2016, 61, 1–15.
- Jung, J.; Hong, H.; Goo, J.M. Ground-glass nodule segmentation in chest CT images using asymmetric multi-phase deformable model and pulmonary vessel removal. Comput. Biol. Med. 2018, 92, 128–138.
- Devi, K.Y.; Sasikala, M. Labeling and clustering-based level set method for automated segmentation of lung tumor stages in CT images. J. Ambient. Intell. Humaniz. Comput. 2021, 12, 2299–2309.
- Shen, W.; Zhou, M.; Yang, F.; Yu, D.; Dong, D.; Yang, C.; Zang, Y.; Tian, J. Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recognit. 2017, 61, 663–673.
- Feng, Y.; Hao, P.; Zhang, P.; Liu, X.; Wu, F.; Wang, H. Supervoxel based weakly-supervised multi-level 3D CNNs for lung nodule detection and segmentation. J. Ambient. Intell. Humaniz. Comput. 2019.
- Wang, S.; Zhou, M.; Gevaert, O.; Tang, Z.; Dong, D.; Liu, Z.; Tian, J. A multi-view deep convolutional neural networks for lung nodule segmentation. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju Island, Republic of Korea, 11–15 July 2017; pp. 1752–1755.
- Huang, X.; Sun, W.; Tseng, T.L.B.; Li, C.; Qian, W. Fast and fully-automated detection and segmentation of pulmonary nodules in thoracic CT scans using deep convolutional neural networks. Comput. Med. Imaging Graph. 2019, 74, 25–36.
- Kumar, S.; Raman, S. Lung nodule segmentation using 3-dimensional convolutional neural networks. In Proceedings of the Soft Computing for Problem Solving: SocProS 2018, Vellore, India, 17–19 December 2018; Springer: Berling, Germany, 2020; Volume 1, pp. 585–596.
- Li, R.; Xiao, C.; Huang, Y.; Hassan, H.; Huang, B. Deep learning applications in computed tomography images for pulmonary nodule detection and diagnosis: A review. Diagnostics 2022, 12, 298.
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017, 35, 18–31.
- Sun, W.; Zheng, B.; Qian, W. Automatic feature learning using multichannel ROI based on deep structured algorithms for computerized lung cancer diagnosis. Comput. Biol. Med. 2017, 89, 530–539.
- Zhao, X.; Sun, W.; Qian, W.; Qi, S.; Sun, J.; Zhang, B.; Yang, Z. Fine-grained lung nodule segmentation with pyramid deconvolutional neural network. In Proceedings of the Medical Imaging 2019: Computer-Aided Diagnosis. International Society for Optics and Photonics, San Diego, CA, USA, 16–21 February 2019; Volume 10950, p. 109503S.
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440.
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; Springer: Berling, Germany, 2016; pp. 424–432.
- Wang, S.; Zhou, M.; Liu, Z.; Liu, Z.; Gu, D.; Zang, Y.; Dong, D.; Gevaert, O.; Tian, J. Central focused convolutional neural networks: Developing a data-driven model for lung nodule segmentation. Med. Image Anal. 2017, 40, 172–183.
- Cao, H.; Liu, H.; Song, E.; Hung, C.C.; Ma, G.; Xu, X.; Jin, R.; Lu, J. Dual-branch residual network for lung nodule segmentation. Appl. Soft Comput. 2020, 86, 105934.
- Rocha, J.; Cunha, A.; Mendonça, A.M. Conventional Filtering Versus U-Net Based Models for Pulmonary Nodule Segmentation in CT Images. J. Med. Syst. 2020, 44, 1–8.
- Singadkar, G.; Mahajan, A.; Thakur, M.; Talbar, S. Deep Deconvolutional Residual Network Based Automatic Lung Nodule Segmentation. J. Digit. Imaging 2020, 33, 678–684.
- Chen, W.F.; Ou, H.Y.; Lin, H.Y.; Wei, C.P.; Liao, C.C.; Cheng, Y.F.; Pan, C.T. Development of Novel Residual-Dense-Attention (RDA) U-Net Network Architecture for Hepatocellular Carcinoma Segmentation. Diagnostics 2022, 12, 1916.
More
