The RAS-ERK pathway is a fundamental signaling cascade crucial for many biological processes including proliferation, cell cycle control, growth, and survival; common across all cell types. Notably, ERK1/2 are implicated in specific processes in a context-dependent manner as in stem cells and pancreatic β-cells. Alterations in the different components of this cascade result in dysregulation of the effector kinases ERK1/2 which communicate with hundreds of substrates. Aberrant activation of the pathway contributes to a range of disorders, including cancer.
- ERK1/2
- MAPKs
- cancer
- scaffold
- therapies
- inhibitors
1. MAPK Pathways
2. ERK Pathway Components
2.1. RAS
RAS (RAt Sarcoma) refers to small GTPase (G)-proteins encoded by three genes yielding proteins with molecular weights of ~21 kDa. RAS proteins contain a GTP-binding domain and a hypervariable region (HVR) which marks the following differences between them [13]: HRAS (Harvey sarcoma viral oncogene), NRAS (neuroblastoma oncogene) and KRAS (Kirsten sarcoma viral oncogene). KRAS has two alternatively spliced forms named KRAS4A and KRAS4B. To be active, RAS proteins need to be localized at the plasma membrane or endomembranes. Lipid post-translational modifications (PTMs), farnesylation and palmitoylation, are essential for RAS association on the membrane [14,15][14][15]. The membrane sub-domain and subcellular localization from which the RAS signal originates determines the downstream effectors that are activated and, consequently, the cellular outcome [16]. As GTPases, RAS proteins cycle from the GDP-loaded OFF configurations to GTP-loaded ON conformations assisted by RAS–guanine nucleotide exchange factors (RAS-GEFs) and return to OFF states facilitated by RAS–GTPase-activating proteins (RAS-GAPs) [17]. The RAS-GEFs SOS (Son of Sevenless)1 and SOS2 are activated by receptor tyrosine kinases coordinated by the adaptor protein Grb2 [18]. Once activated, RAS can interact with the RAS binding domain (RBD) in RAS effectors. In addition to RAF proteins, MAP3Ks in the ERK1/2 pathway, there are over fifty other RAS effectors including RALGDS, RALBP1, RIN1, TIAM1, PLCε, REPAC, RASSF1/5, and PI3K among others [19,20][19][20]. The four RAS proteins show around 85% of amino acid identity differing in the hypervariable region (HVR) [21]. In spite of their similarities, manipulation of their expression or activation results in different cellular outcomes [22,23][22][23]. KRAS is the only family member essential for development [24,25,26][24][25][26]. This feature depends on its locus expression pattern in development, as the protein product of HRAS knock-in in the KRAS locus gives rise to viable mice. However, a phenotype in adult life has been observed when KRAS is replaced by HRAS [27], although it disappears by eliminating the endogenous HRAS [28]. These findings indicate that different RAS isoforms are apparently equivalent in physiological conditions. Despite the high amino acid identity between RAS isoforms, their nucleotide sequences are less so [31][29]. The Counter laboratory proposed codon bias as a mechanism responsible for the different expression and oncogenicity of KRAS and HRAS. Even though KRAS is the most commonly mutated isoform, it is the most poorly translated due to rare codons. This group suggested that the potent oncogenicity of HRAS lead to cell cycle arrest and senescence, while the different codon usage of KRAS overcomes this issue becoming the most frequently mutated RAS in cancer [31][29]. In fact, they later demonstrated that substitution of KRAS rare codons for common codons rendered tumors less aggressive [32][30]. Following studies corroborated that rare codons in KRAS affect transcription due to histone modification and transcriptional activation recruitment, mRNA translation, and even protein conformation, highlighting the importance of codon usage [33][31].2.2. RAF
RAF (Rapidly Accelerated Fibrosarcoma) is the first kinase in the pathway core, serving as the MAP3K downstream of RAS [39][32]. The regulatory paradigm has it that RAF is cytosolic in resting conditions and translocates to the plasma membrane to be activated by RAS upon growth factor stimulation [40,41,42][33][34][35] (reviewed in [43,44][36][37]). In mammals, the RAF family consists of ARAF (65 kDa), CRAF (65 kDa) (aka RAF1), and BRAF (84 kDa, larger due to additional BRAF-specific sequence at its N-terminus) [45][38], which are ~75% identical in shared segments. Also in the family are the pseudokinases KSR1 and KSR2 discussed in Section 4.2. RAF consists of a RAS binding domain (RBD) and a cysteine-rich domain (CRD), both in the N-terminal regulatory region of the protein and the kinase domain in the C-terminus. RAF was formerly divided into three conserved regions named CR1, CR2, and CR3. CR1 contains the RBD and the CRD, CR2 is a serine/threonine-rich domain, and CR3 refers to the kinase domain (reviewed in [46][39]). CRD facilitates RAF localization at the plasma membrane [47,48][40][41] and together with RBD both interact with RAS [49,50][42][43]. Of the three isoforms, BRAF is the most frequently mutated in cancer. BRAF mutations are present in 7.7% across all cancer types [60][44]. Overall, BRAF mutation prevalence in human cancers is almost 100% for hairy cell leukemia, 50% for melanoma, 45% for papillary thyroid cancer, 10% for colon cancer, and 10% for non-small cell lung cancer cases [61][45]. The most frequent mutation in BRAF is V to E substitution at residue 600 (V600E), accounting for 90% of BRAF mutations [62][46]. Around 200 low-frequency cancer-associated mutations have been described in BRAF [63][47]. Most of them are believed to reverse the autoinhibitory conformation in the absence of upstream signaling [64][48]. BRAF mutations have been classified into three groups as follows: class I mutations are those that affect BRAF V600 position, most commonly V600E but also V600K/D/R/M which are able to transduce the signal downstream as monomers; class II mutations function as RAS-independent dimers with reported increased kinase activity but still weaker than BRAF V600 mutants; and class III mutants show impaired kinase activity and do not directly phosphorylate MEK but retain the ability to bind RAS and heterodimerize with CRAF [65,66][49][50].2.3. MEK
Downstream of RAF are the two ERK-specific MAP2Ks [68[51][52],69], MEKs 1 and 2 of 44 and 45 kDa, respectively [70][53]. The MEK kinase domain contains an ERK-selective binding site near the N-terminus and a nuclear export sequence (NES) that favors its cytosolic localization [71,72][54][55]. MEK1 is activated by phosphorylation of two activation loop serine residues (S218 and S222, S222 and S226 in MEK2) [73][56]. Apart from canonical activation by RAF, MEK can also be activated by other MAP3Ks, including MOS [74][57], MAP3K1/MEKK1 [75][58], MAP3K8 (aka Tpl2/Cot) [76][59], and MAST1 [77][60], linking activation to distinct upstream stimuli (Figure 1).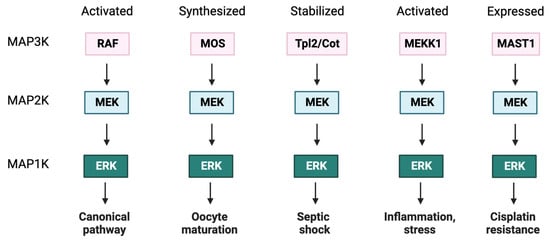
2.4. ERK
2.4.1. ERK Substrate Recognition
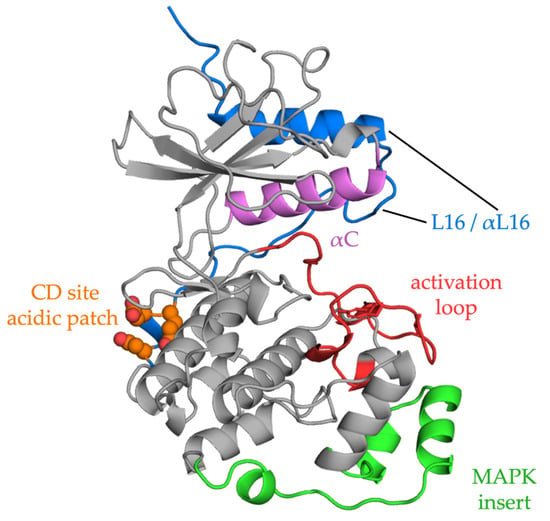
ERKs can interact with hundreds of substrates in any location in the cell [119,120][85][86]. In addition to the many phosphorylation sites that have been identified by examining defined targets, many more sites in putative effectors have been predicted by in silico approaches with some validated in vivo [121,122][87][88]. ERK1/2 show a preference for sites in the consensus phosphorylation sequence, PX(S/T)P, with a proline in the P+1 residue, following the phosphorylation site, and are thus considered proline-directed kinases. Many substrates contain only a minimal (S/T)P sequence. The huge number of phospho-sites found in proteins [121][87] raises the question of how specificity is achieved. While scaffolds can form three-way complexes with enzymes and substrates, docking sites are important MAPK recognition mechanisms [124,125][89][90]. The best characterized docking sites identified on ERK are the common docking (CD) or D-recruitment site (DRS) and the F-recruitment site (FRS) or docking site for ERK FXF (DEF) (see Figure 2) [126][91]. The CD site, accessible in both inactive and active ERK configurations, binds a basic/hydrophobic motif, typically K/RX2-4LXL, referred to as a D motif, kinase interaction motif (KIM), or more generally as a short linear motif (SLiM). In addition to many substrates, MEK1/2 also bind ERK1/2 through a CD–D motif interaction, as does the MAP kinase phosphatase DUSP6. The FRS site binds proteins containing FXF motifs, including nuclear pore proteins [127][92], and is more accessible after ERK activation [128,129][93][94].2.4.2. ERK Localization
ERK1/2 are found in cytoplasmic-, nuclear-, and membrane-associated compartments, and their subcellular distribution defines access to substrates. Localization is dynamic and is influenced by the duration and nature of activating stimulation. Two ERK monomers can associate to form a free dimer in the cytoplasm to activate cytoplasmic substrates. ERK dimers can be assembled on scaffold proteins in specific subcellular localizations where the ERK monomer not associated with the scaffold will phosphorylate specific substrates [137][95]. Binding the FG repeat regions of nuclear pore proteins, nucleoporins (Nup) Nup214, Nup153, TPR, and others, allows passive entry of ERK into the nucleus [138,139,140][96][97][98]. While active nuclear import mechanisms have been identified, ERK scaffolding, to retain the kinases in the relevant compartment, and ERK nuclear export through binding to exported partners, appear to dominate processes determining ERK location. As noted earlier, binding to MEK promotes ERK nuclear export and favors cytoplasmic retention. Dual specificity phosphatases also influence ERK location by related mechanisms. The phosphatase DUSP6 is primarily cytoplasmic and binds ERK through a D motif–CD domain interaction; DUSP6 also possesses an NES, promoting ERK nuclear export.2.4.3. ERK in Cancer
In contrast to upstream cascade components, ERK1/2 are rarely mutated and oncogenic activity has not been unequivocally demonstrated. ERK mutations recognized as of 2020 have been thoroughly reviewed by Smorodinsky-Atias (2020) [151][99]. Most of these mutations have been generated in the laboratory based on ERK orthologs, protein structure and two ancestors as follows: one common to ERK1/2/5 with autophosphorylation capabilities and another common to ERK1 and ERK2 that lacks the autophosphorylation ability. A few of them rendered some activation but usually minimal compared to that achieved by MEK [152][100]. An ERK mutation with potential oncogenic activities is an intrinsically active mutant ERK1 with the mutation R84S, initially found in a screen for ERK MEK-independent mutants in an ERK yeast ortholog [154][101]. This mutant was able to transform NIH3T3 cells [155][102] and was activated in developmental assays in a Drosophila cancer model, especially combined with the sevenmaker mutation [156][103]. However, the equivalent mutation in ERK2 did not replicate the transformation outcomes. According to the COSMIC database, as of September 2023, ERK1 accounts for 0.5% of all the cancer samples analyzed. These mutations can be found throughout the protein with no hotspots identified, most of them were found only in one single sample with a maximum of four cases [157][104]. Mutations in ERK2 are slightly more frequent, representing 1.2% of the tested samples. Alterations also spread across the whole protein; however, there is a hotspot in residues 321 and 322 within the common docking domain. In fact, the most common mutation is E322K, which occurs in only 0.1% of all the assessed samples but accounts for 7% of all ERK2 mutations found in cancer samples. Although the majority of the mutations in ERK1 and ERK2 do not follow a tissue distribution pattern, mutations in the CD site of ERK2 are found mainly in squamous cell carcinoma, most frequently in cervix, oral-esophagus, head and neck, lung and oral tissues [159][105]. E322K was originally identified in oral squamous cell carcinoma [160][106].3. ERK Functions and Context Specificity
ERK1/2 are critical for integrating extracellular ligand binding cues with intracellular conditions to elicit coordinated cellular responses such as exit from a stem cell state, proliferation, terminal differentiation, and tissue-specific differentiated functions [165,166][107][108]. As noted above, ERK2 is essential early in embryonic development, and variation in a single allele is associated with developmental and cognitive impairment [104,167,168,169][82][109][110][111]. ERK2 can phosphorylate hundreds of downstream targets, including more than half a dozen protein kinase substrates which expand its regulatory reach in many subcellular locations [122,141,170,171,172,173][88][112][113][114][115][116]. The ability to communicate directly and indirectly with such a breadth and distribution of downstream proteins results in both the intrinsic potential of ERK2 to carry out ligand-specific directives and also the malleability of ERK2-elicited responses to context, tightly linked to the cellular proteome expressed at any specific time [174,175,176,177,178][117][118][119][120][121]. This plasticity also makes cells vulnerable to diseases arising from disturbances in ERK2 regulation. The activities of ERK1/2 must be adjusted up and down during the cell cycle, as development progresses, and in cells of the adult animal. This fluctuation in ERK1/2 activity is required to maintain or exit from a stem cell state, to facilitate terminal differentiation, and to maintain cell identity. ERK1/2 activities are necessary participants that mediate acute endocrine, paracrine, and autocrine stimulation, and adjust outputs due to changes in nutrient availability and other cellular stresses to support the context-specified cellular responses.3.1. Cell Cycle, Growth, Migration, and Survival
ERK regulates G1 to S phase transition, and its depletion results in cell cycle arrest [179][122] (reviewed in [180][123]). Cell growth is essentially biomass accumulation due to anabolic growth pathways [181][124]. ERK participates in the regulation of several processes implicated in growth for the preparation of cell division (reviewed in [182][125]). For instance, ERK stimulates the de novo synthesis of pyrimidine nucleotides to generate DNA and RNA. ERK phosphorylates the CAD protein (carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase), increasing pyrimidine biosynthesis and antagonizing the negative effect of PKA CAD phosphorylation [183][126]. Ribosomes are necessary to handle new protein synthesis. ERK stimulates ribosome biogenesis by phosphorylation of RNA polymerase I [184][127] and III [185][128]. Finally, ERK is involved in protein translation by regulation of mammalian target of rapamycin complex 1 (mTORC1), eukaryotic translation initiation factor 4E (eIF4E) through MNK1/2, and cytoplasmic polyadenylation element binding protein 1 (CPEB1) [186][129] (reviewed in [187][130]). ERK increases mRNA polyadenylation and translation [188][131].3.2. Context Dependence of ERK Functions in Pancreatic Beta Cells
In pancreatic beta cells, circulating glucose regulates insulin secretion and production. ERK1/2 are essential in nutrient sensing, and their activities rise and fall as a function of glucose concentration over the physiologic range, in parallel with insulin secretion [203][132]. Glucose metabolism triggers calcium influx and release from intracellular stores to activate ERK1/2. Calcium influx also activates the calcium-dependent phosphatase calcineurin, which is required for maximal ERK1/2 activation by glucose [204][133]. Calcineurin is required for activation of ERK1/2 by other stimuli that induce insulin secretion as well, including depolarization and glucagon-like peptide 1, but not by phorbol ester or insulin. BRAF is also a calcineurin substrate; calcineurin dephosphorylates threonine 401 on BRAF, which is a site of negative feedback phosphorylation by ERK1/2 [205][134]. The major calcineurin-dependent event in glucose sensing by ERK1/2 is the activation of BRAF in beta cells [206][135].3.3. ERK in Stemness
The requirement for ERK in stem cells depends on the pluripotency state of the cells. There are two states of pluripotency as follows: naïve, that resembles an early-stage embryo (pre-implantation) and primed stem cells, that evoke a later stage (post-implantation) [216][136]. Naïve pluripotent cells require LIF/STAT3 (leukemia inhibitory factor/signal transducer and activator of transcription 3) signaling, whereas primed pluripotency depends on FGF (fibroblast growth factor)/ERK and activin signaling [217][137]. ERK activation is dispensable for naïve but essential for primed pluripotency. In the case of mouse embryonic stem cells, the activation of the LIF/STAT3 pathway [218,219][138][139] and BMP (bone morphogenetic protein) triggering Id (inhibition of differentiation) genes are enough to maintain pluripotency. To avoid differentiation, they are cultured in the presence of LIF [218][138] and BMP4 [220][140]. In this setting, ERK antagonizes STAT3 signaling promoting differentiation [221][141], but BMP4 suppresses ERK signaling to sustain self-renewal through Smad activation and DUSP9 upregulation [222,223][142][143]. Meanwhile, GSK3 (glycogen synthase kinase-3) inhibits Wnt/β-catenin signaling, which is essential for maintaining self-renewal and inhibiting differentiation in mESCs [224,225][144][145]. mESCs require MEK and GSK3 inhibitors in the culture medium for stability; this combination is known as 2i-culture [226,227][146][147]. ERK activation is crucial in mESCs to end self-renewal and initiate differentiation [228,229][148][149]. Stem cells from mice (mESCs) and human (hESCs) do not behave identically. mESCs are derived from pre-implantation embryos [231[150][151],232], while hESCs are acquired from post-implantation embryos. Thus, mESCs display naïve pluripotency [233,234[152][153][154],235], whereas hESCs belong to a primed pluripotent state [236][155].3.4. ERK as an Allosteric Regulator
ERK possesses multiple functions independent of its catalytic activity. In addition to the many activity-dependent actions on transcription factors (e.g., Elk1, NeuroD1) noted above, ERK also influences gene expression transcription factors, co-activators, and co-repressors through direct interactions. For instance, ERK acts as a transcriptional repressor for interferon gamma (IFNγ)-induced genes as both ERK2 and C/EBP-β compete to bind GATE element, and only when C/EBP-β is bound are the downstream genes expressed [244][156]. ERK directly assembles on a promoter element of the IL4 gene, aiding in recruitment of transcription factors that initiate IL4 gene transcription. Association with the promoter and IL4 transcription induction is necessary for human Th2-cell differentiation [245][157]. ERK2 directly binds the MYC promoter and recruits CDK9 which binds and activates RNA polymerase II, resulting in ERK-induced MYC expression independently of ERK activity [246][158]. ERK allosterically regulates certain other proteins, notably DUSP6, independent of its kinase activity. A D motif in the non-catalytic amino-terminal domain of DUSP6 binds the ERK CD site. This interaction stabilizes the active conformation of DUSP6 by inducing the closure of the general acid loop in the phosphatase [248][159]. Thus, ERK provokes a conformational change in the DUSP6 catalytic domain increasing its phosphatase activity to negatively regulate ERK pathway activation [249][160].4. ERK Regulation
4.1. ERK Negative Feedback Loops
Among its extensive substrates, ERK can bind and phosphorylate upstream members of the cascade to exert a positive or negative impact on the downstream signal. Negative feedback loops downregulate the signaling maintaining pathway balance. In addition to upstream components, ERK can phosphorylate scaffolds and adaptor proteins to rapidly suppress pathway activity, referred to as direct regulation. ERK can also phosphorylate transcription factors responsible for the expression of negative regulators, giving rise to a delayed but long-term response known as indirect regulation (reviewed in [254][161]). ERK can phosphorylate multiple sites on BRAF and CRAF, some preventing their heterodimerization and others the interaction with RAS and localization at the plasma membrane [255,256][162][163] (Figure 3). In the case of BRAF, ERK also phosphorylates several residues with a negative outcome [205][134]. The conserved motif SPKTP, not present in the other RAF isoforms, is found in the C-terminus of BRAF, and it is also phosphorylated by ERK and implicated in the negative regulation of the pathway [257][164]. Three other residues in BRAF, T401, S750 and T753, are phosphorylated by ERK and prevent heterodimerization with CRAF [205,258][134][165].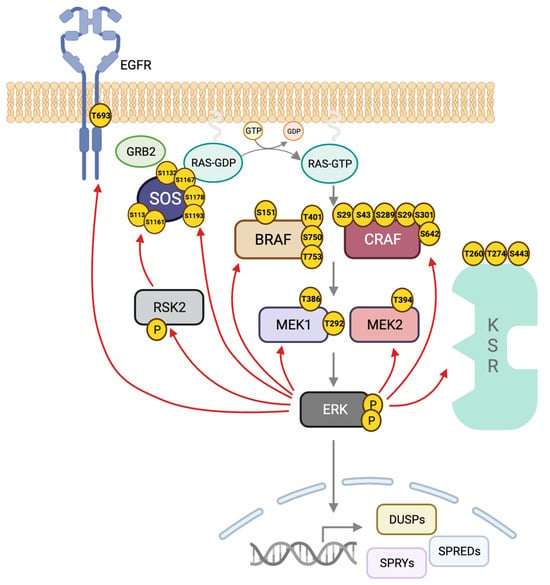
4.2. ERK Scaffolds
4.2.1. KSR1/2
In 1995, KSR1 was simultaneously discovered in Drosophila virilis/Drosophila melanogaster [333][191] and Caenorhabditis elegans [334,335][192][193] by a genetic screen for RAS downstream effectors. The phenotypes of the KSR1 loss-of-function mutants revealed inhibition of RAS signals, and for this reason the protein was named Kinase Suppressor of RAS (KSR) [333,334][191][192]. KSR was recognized as a mammalian functional equivalent of the yeast MAPK scaffold protein Ste5, even though they do not share sequence homology. The KSR family is composed of five members, the pseudokinases KSR1 [333][191] and KSR2 [336][194] and their relatives, the three RAF protein kinases. Despite close sequence similarity between KSR1 and KSR2, these two proteins have distinct functions which are due in part to differences in expression. While KSR1 is widely expressed, KSR2 has a restricted tissue expression pattern, generally with neuroendocrine enrichment. Regarding function, both are scaffolds of the RAS–ERK pathway, but the knockout phenotypes are markedly different. Mice lacking KSR1 show a mild phenotype with disorganized hair follicles but no developmental issues, while RAS-directed oncogenesis is reduced [323][181]. On the other hand, KSR2-deficient mice are less fertile and develop metabolic disorders such as obesity, insulin resistance and type 2 diabetes [337,338][195][196]. Why are KSR1/2 considered pseudokinases? Although mammalian KSR1/2 were initially classified as pseudokinases because they contain arginine in place of the lysine essential for catalytic activity, this has been a point of contention [348,349,350,351,352,353][197][198][199][200][201][202]. Some studies have reported residual kinase activity [306,351,354,355][200][203][204][205] not detected by others [342,356,357][206][207][208]. Indirect evidence suggesting catalytic function comes from the findings that ectopic expression of KSR1 or KSR2 is sufficient to induce proliferation in a RAS-independent manner, attributed to heterodimerization with RAF. Still, the exact mechanism by which KSR overexpression leads to ERK activation has not been defined [358][209]. KSR1 is usually cytoplasmic and thought to be constitutively bound to MEK [345,346,347,359][210][211][212][213]. Upon stimulation and RAS activation, KSR1 rapidly translocates to the plasma membrane where it interacts with a RAF isoform, especially with BRAF [58][214]. At the plasma membrane, KSR exhibits selectivity towards defined microdomains, responding preferentially to RAS signals from lipid rafts [339][215]. Interaction with the E3 ligase IMP (Impedes Mitogenic signal Propagation) promotes the recruitment of KSR to Triton-resistant structures that sequester KSR1 and block ERK activation [360,361][216][217]. RAS activation leads to IMP proteasomal degradation, facilitating KSR-mediated ERK activation [362][218]. Homo- and heterodimerization of KSR and RAF proteins underscores the remarkable complexity of ERK regulation at the MAP3K level, some of which was originally uncovered through studies of RAF inhibitors. A depth of structural data has revealed multiple dimeric structures of these molecules [57,306][203][219]. KSR and RAF can form side-to-side heterodimers believed to trigger RAF activation through a nearly identical dimer interface that is conserved across all family members. In the same way, KSR1 can also homodimerize through its C-terminus, forming side-to-side dimers as well [57,367][219][220]. Apart from KSR–RAF interaction through the kinase domains, selective heterodimerization of RAF with KSR1 occurs through direct contacts between the N-terminal regulatory regions of each protein, including the KSR coiled-coil–sterile alpha motif (CCSAM) [340][221].4.2.2. IQGAP1/2 and 3
IQ motif-containing GTPase Activating Protein (IQGAP) 1 was first identified in 1994 by Weissbach and collaborators in a screen to discover novel matrix metalloproteases [368][222]. Nevertheless, it was not until 2004 that IQGAP was classified as a Ras/ERK pathway scaffold [324][182]. Mammalian IQGAP1, 2, and 3 are multidomain proteins of ~190 kDa [369,370,371][223][224][225]. IQGAPs exhibit different tissue expression patterns; while IQGAP1 is ubiquitous [368][222], IQGAP2 is mainly expressed in the liver and the gastro-intestinal and urogenital track [372[226][227],373], and IQGAP3 is found mainly in the brain, lung and testes [374][228]. IQGAP1 is cytoplasmic and mainly associated with the cytoskeleton, with particular enrichment in cell–cell contacts [375][229]. Distributions of IQGAP2 and IQGAP3 are not well described, but they have been noted throughout the cytoplasm and in cell–cell junctions, respectively [376,377][230][231]. IQGAP isoforms share a similar domain composition (Figure 54). Abundant protein-interacting motifs make them key players in numerous cellular processes. For example, more than 100 binding partners have been described for IQGAP1 [379][232]. At the N-termini of IQGAPs is a calponin homology domain (CHD) that binds cytoskeletal proteins, in particular F-actin [380][233]. This domain can also bind calmodulin/Ca2+ but at a lower affinity than to the IQ domain and in competition with F-actin [381][234]. The CHD is followed by a WW domain with two conserved Trp residues (W). WW domains specify proline-rich regions in binding partners. Interestingly, a major IQGAP WW domain-binding partner, ERK, apparently binds through a different mechanism [324][182]. Four tandem isoleucine/glutamine-containing (IQ) motifs are multipurpose domains that bind partners including calmodulin [381,382][234][235], the EGF receptor [383][236], MEK [384][237], BRAF [385][238] and the small GTPase RAP1 [386][239]. IQ domains are also important as they mediate the formation of IQGAP dimers and oligomers [387][240]. IQ domains participate in the interaction with phosphoinositide signaling elements such as phosphatidylinositol phosphate kinase type Iγ (PIPKIγ) at the leading edge. PIPKIγ binding favors the IQGAP open conformation to recruit the actin polymerization machinery and participate in cell motility [388][241].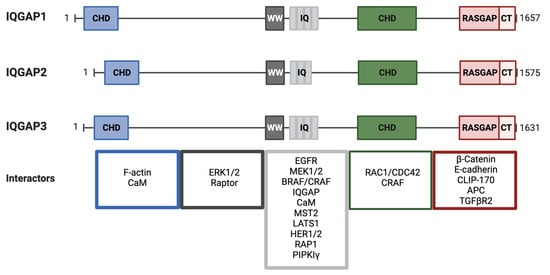
4.2.3. HPIP
5. Pathway Inhibitors
5.1. Classical Inhibitors
Many classical inhibitors for the different members of the cascade have been developed. Many of them are in clinical trials but only a few have received FDA approval, mostly those targeting RAF and MEK. Remarkably, two covalent inhibitors, sotorasib and adragasib, have been FDA-approved in the last couple of years to treat KRASG12C lung cancers (reviewed in [487][254]), and several promising clinical trials are currently in progress [30][255]. As of today, no ERK inhibitors are in clinical use, although several are in clinical trials [488][256]. In contrast to many other kinases, ERK2 undergoes relatively small conformational changes in the active site upon activation. Solution measurements have found evidence for phosphorylated ERK2 (2P-ERK2) in two conformational states, R and L, which interconvert on a millisecond time scale [490][257]. Surprisingly, the L state resembles the active site of unphosphorylated ERK (0P-ERK2). A recent study found ERK inhibitors that interact with both states and ones that selectively interact with the R or L states. Inhibitors were noted that could shift the equilibrium towards the R or L states. As some ERK inhibitors have been shown to affect specific protein–protein interactions, this study provides new insights for inhibitor design [491][258].5.2. PROTAC Technology Applied to the ERK Pathway
As its name implies, Targeted Protein Degradation (TPD) covers several methods to induce the selective degradation of a specific molecule either via the proteasome, lysosome or autophagy system. One of these methods using the proteasome degradation system is PROTAC (Proteolysis-Targeting Chimera) [514][259], initially developed in 2001 [515,516,517,518][260][261][262][263]. The strategy involves recruitment of an E3 ligase, usually VHL (von Hippel-Lindau VHL) or CRBN (Cereblon), in current versions of PROTACs, to the target protein to tag it for degradation. Two ligands, one that binds the target protein and a second that recruits an E3 ligase, are connected through a chemical linker. The target protein will be ubiquitinylated by the E3 and sent to the proteasome resulting in its near elimination [519][264]. Advantages of protein degradation versus classical small molecule inhibitors are many. PROTACs can reach the undruggable proteome as long as a suitable target ligand is available. Degradation prevents target protein accumulation, thus avoiding compensatory upregulation and drug resistance. PROTACs overcome the generation of resistant mutations under selective pressures or even the emergence of non-enzymatic functions of the targeted proteins, which might provoke a signaling rebound. The major limitation of PROTACs continues to be poor oral bioavailability and low cell permeability due to their large sizes [523][265]. For this reason, it was not until 2019 that the first PROTAC progressed to clinical trials [524][266]. Although none are employed in the RAS–ERK pathway, a few FDA-approved therapies use degraders [525][267]. The first attempt to generate a PROTAC against KRASG12C was based on the inhibitor ARS1620, and a thalidomide analog, pomalidomide, an FDA-approved drug with high affinity for the ubiquitin ligase CRBN. The compound was named XY-4-88, but failed to induce endogenous KRASG12C degradation in cancer cells [526][268]. A parallel study generated LC-2, the first successful PROTAC against KRASG12C, which used MRTX849 as the parental inhibitor [527][269]. The first series of compounds acting as PROTACs for BRAF were created based on the inhibitor rigosertib, which binds to RAF in the RAS binding domain (RBD) and should, therefore, bind RAF mutants as activating mutations are in the kinase domain [529][270], although this was not tested. P4B was the first effective PROTAC with selectivity for the BRAF mutant V600E. The ligand was based on the inhibitor BI882370 and provides degradative efficacy in the nanomolar range [530][271]. The first-in-class highly selective degrader of MEK1 and MEK2 was the PD0325901-based compound MS432. It was effective in colorectal cancer and melanoma cell proliferation, with good bioavailability in mice [534][272]. A few PROTAC compounds against MEK1 and MEK2 were developed based on allosteric MEK inhibitors, such as PD0325901 and refametinib, using VHL as the E3 ligase. They were assessed in cell proliferation and pERK levels to compare the compound with the parental inhibitor. Although these compounds had modest degradation efficiencies, two of them were able to completely suppress proliferation in melanoma cells [535][273]. Following the development of the first-in-class MEK degrader, the Lin laboratory also characterized three more, MS928, MS934 and MS910. Compared with previously reported VHL-recruiting degraders, these new compounds were more potent in preserving the high selectivity. Of these three degraders, MS934 appeared as the best MEK1/2 degrader for in vivo studies to date.5.3. Molecular Glues Targeting MEK
The natural products cyclosporin A and FK506 were among the earliest described as molecular glues by Schreiber in 1992 for their ability to promote interactions between two molecules that do not otherwise bind. Following this molecular glue concept, Simonetta (2019) [543][274] set out to develop a compound that would enhance a protein–protein interaction between a transcription factor and the E3 that normally degrades it. Subsequently, more molecular glues have been identified by high-throughput chemical screens and target validation [544,545][275][276]. Because molecular glues are small molecules, unlike PROTACs, they should have superior membrane penetrability and oral bioavailability. On the other hand, PROTACs display a higher adaptability to target a protein of interest, while the original molecular glues had unknown ligands [523,546][265][277].5.4. Optically Activated MEK Inhibitors
Light-dependent pharmacological approaches, specifically photo-decaging, have been proposed to prevent MEKi toxicities. The strategy consists of generating an inactive locked MEK inhibitor precursor, by linking a photo-cleavable protecting group (the ‘cage’) to the small molecule inhibitor. This drug protector is irreversibly cleaved from the MEK inhibitor by exposure to light, and then the inhibitor will bind MEK where it has been uncaged. This cutting-edge strategy has been carried out with the potent allosteric inhibitor PD0325901, which was previously discontinued in clinical trials due to its harmful effects which are presumably on-target. The optically activatable MEK1/2 inhibitors, named opti-MEKi, have shown promising efficacy in vivo in a melanoma xenograft zebrafish model [550][278]. Combinations of these approaches, e.g., photo-switchable linkers or light-activated degraders, are also in development [551][279].
References
- Cooper, J.A.; Bowen-Pope, D.F.; Raines, E.; Ross, R.; Hunter, T. Similar Effects of Platelet-Derived Growth Factor and Epidermal Growth Factor on the Phosphorylation of Tyrosine in Cellular Proteins. Cell 1982, 31, 263–273.
- Ray, L.B.; Sturgill, T.W. Characterization of Insulin-Stimulated Microtubule-Associated Protein Kinase. Rapid Isolation and Stabilization of a Novel Serine/Threonine Kinase from 3T3-L1 Cells. J. Biol. Chem. 1988, 263, 12721–12727.
- Sturgill, T.W.; Ray, L.B.; Erikson, E.; Maller, J.L. Insulin-Stimulated MAP-2 Kinase Phosphorylates and Activates Ribosomal Protein S6 Kinase II. Nature 1988, 334, 715–718.
- Boulton, T.G.; Yancopoulos, G.D.; Gregory, J.S.; Slaughter, C.; Moomaw, C.; Hsu, J.; Cobb, M.H. An Insulin-Stimulated Protein Kinase Similar to Yeast Kinases Involved in Cell Cycle Control. Science 1990, 249, 64–67.
- Boulton, T.G.; Cobb, M.H. Identification of Multiple Extracellular Signal-Regulated Kinases (ERKs) with Antipeptide Antibodies. Mol. Biol. Cell 1991, 2, 357–371.
- Boulton, T.G.; Gregory, J.S.; Cobb, M.H. Purification and Properties of Extracellular Signal-Regulated Kinase 1, an Insulin-Stimulated Microtubule-Associated Protein 2 Kinase. Biochemistry 1991, 30, 278–286.
- English, J.; Pearson, G.; Wilsbacher, J.; Swantek, J.; Karandikar, M.; Xu, S.; Cobb, M.H. New Insights into the Control of MAP Kinase Pathways. Exp. Cell Res. 1999, 253, 255–270.
- Coulombe, P.; Meloche, S. Atypical Mitogen-Activated Protein Kinases: Structure, Regulation and Functions. Biochim. Biophys. Acta-Mol. Cell Res. 2007, 1773, 1376–1387.
- Cargnello, M.; Roux, P.P. Activation and Function of the MAPKs and Their Substrates, the MAPK-Activated Protein Kinases. Microbiol. Mol. Biol. Rev. 2011, 75, 50–83.
- Chartier, M.; Chénard, T.; Barker, J.; Najmanovich, R. Kinome Render: A Stand-Alone and Web-Accessible Tool to Annotate the Human Protein Kinome Tree. PeerJ 2013, 1, e126.
- Robbins, D.J.; Zhen, E.; Cheng, M.; Xu, S.; Vanderbilt, C.A.; Ebert, D.; Garcia, C.; Dang, A.; Cobb, M.H. Regulation and Properties of Extracellular Signal-Regulated Protein Kinases 1, 2, and 3. J. Am. Soc. Nephrol. 1993, 4, 1104–1110.
- Pearson, G.; Robinson, F.; Gibson, T.B.; Xu, B.E.; Karandikar, M.; Berman, K.; Cobb, M.H. Mitogen-Activated Protein (MAP) Kinase Pathways: Regulation and Physiological Functions. Endocr. Rev. 2001, 22, 153–183.
- Cox, A.D.; Fesik, S.W.; Kimmelman, A.C.; Luo, J.; Der, C.J. Drugging the Undruggable RAS: Mission Possible? Nat. Rev. Drug Discov. 2014, 13, 828–851.
- Arozarena, I.; Calvo, F.; Crespo, P. Ras, an Actor on Many Stages: Posttranslational Modifications, Localization, and Site-Specified Events. Genes Cancer 2011, 2, 182–194.
- Prior, A.I.; Hancock, J.F. Ras Trafficking, Localization and Compartmentalized Signalling. Semin. Cell Dev. Biol. 2012, 32, 145–153.
- Casar, B.; Pinto, A.; Crespo, P. ERK Dimers and Scaffold Proteins: Unexpected Partners for a Forgotten (Cytoplasmic) Task. Cell Cycle 2009, 8, 1007–1013.
- Hall, B.E.; Bar-Sagi, D.; Nassar, N. The Structural Basis for the Transition from Ras-GTP to Ras-GDP. Proc. Natl. Acad. Sci. USA 2002, 99, 12138–12142.
- Egan, S.E.; Giddings, B.W.; Brooks, M.W.; Buday, L.; Sizeland, A.M.; Weinberg, R.A. Association of Sos Ras Exchange Protein with Grb2 Is Implicated in Tyrosine Kinase Signal Transduction and Transformation. Nature 1993, 363, 45–51.
- Kiel, C.; Matallanas, D.; Kolch, W. The Ins and Outs of Ras Effector Complexes. Biomolecules 2021, 11, 236.
- Kolch, W.; Berta, D.; Rosta, E. Dynamic Regulation of RAS and RAS Signaling. Biochem. J. 2023, 480, 1–23.
- Barbacid, M. ras Genes. Annu. Rev. Biochem. 1987, 56, 779–827.
- Ninomiya, Y.; Kato, K.; Takahashi, A.; Ueoka, Y.; Kamikihara, T.; Arima, T.; Matsuda, T.; Kato, H.; Nishida, J.I.; Wake, N. K-Ras and H-Ras Activation Promote Distinct Consequences on Endometrial Cell Survival. Cancer Res. 2004, 64, 2759–2765.
- Voice, J.K.; Klemke, R.L.; Le, A.; Jackson, J.H. Four Human Ras Homologs Differ in Their Abilities to Activate Raf-1, Induce Transformation, and Stimulate Cell Motility. J. Biol. Chem. 1999, 274, 17164–17170.
- Johnson, L.; Greenbaum, D.; Cichowski, K.; Mercer, K.; Murphy, E.; Schmitt, E.; Bronson, R.T.; Umanoff, H.; Edelmann, W.; Kucherlapati, R.; et al. K-Ras Is an Essential Gene in the Mouse with Partial Functional Overlap with N-Ras. Genes Dev. 1997, 11, 2468–2481.
- Koera, K.; Nakamura, K.; Nakao, K.; Miyoshi, J.; Toyoshima, K.; Hatta, T.; Otani, H.; Aiba, A.; Katsuki, M. K-Ras Is Essential for the Development of the Mouse Embryo. Oncogene 1997, 15, 1151–1159.
- Esteban, L.M.; Vicario-Abejón, C.; Fernández-Salguero, P.; Fernández-Medarde, A.; Swaminathan, N.; Yienger, K.; Lopez, E.; Malumbres, M.; McKay, R.; Ward, J.M.; et al. Targeted Genomic Disruption of H-Ras and N-Ras, Individually or in Combination, Reveals the Dispensability of Both Loci for Mouse Growth and Development. Mol. Cell. Biol. 2001, 21, 1444–1452.
- Potenza, N.; Vecchione, C.; Notte, A.; De Rienzo, A.; Rosica, A.; Bauer, L.; Affuso, A.; De Felice, M.; Russo, T.; Poulet, R.; et al. Replacement of K-Ras with H-Ras Supports Normal Embryonic Development despite Inducing Cardiovascular Pathology in Adult Mice. EMBO Rep. 2005, 6, 432–437.
- Drosten, M.; Simon-Carrasco, L.; Hernandez-Porras, I.; Lechuga, C.G.; Blasco, M.T.; Jacob, H.K.C.; Fabbiano, S.; Potenza, N.; Bustelo, X.R.; Guerra, C.; et al. H-Ras and K-Ras Oncoproteins Induce Different Tumor Spectra When Driven by the Same Regulatory Sequences. Cancer Res. 2017, 77, 707–718.
- Lampson, B.L.; Pershing, N.L.K.; Prinz, J.A.; Lacsina, J.R.; Marzluff, W.F.; Nicchitta, C.V.; MacAlpine, D.M.; Counter, C.M. Rare Codons Regulate KRas Oncogenesis. Curr. Biol. 2013, 23, 70–75.
- Pershing, N.L.K.; Lampson, B.L.; Belsky, J.A.; Kaltenbrun, E.; MacAlpine, D.M.; Counter, C.M. Rare Codons Capacitate Kras-Driven de Novo Tumorigenesis. J. Clin. Investig. 2015, 125, 222–233.
- Fu, J.; Dang, Y.; Counter, C.; Liu, Y. Codon Usage Regulates Human KRAS Expression at Both Transcriptional and Translational Levels. J. Biol. Chem. 2018, 293, 17929–17940.
- Rapp, U.R.; Goldsborough, M.D.; Mark, G.E.; Bonner, T.I.; Groffen, J.; Reynolds, F.H.; Stephenson, J.R. Structure and Biological Activity of V-Raf, a Unique Oncogene Transduced by a Retrovirus. Proc. Natl. Acad. Sci. USA 1983, 80, 4218–4222.
- Surve, S.V.; Myers, P.J.; Clayton, S.A.; Watkins, S.C.; Lazzara, M.J.; Sorkina, A. Localization Dynamics of Endogenous Fluorescently Labeled RAF1 in EGF-Stimulated Cells. Mol. Biol. Cell 2019, 30, 506–523.
- Hekman, M.; Hamm, H.; Villar, A.V.; Bader, B.; Kuhlmann, J.; Nickel, J.; Rapp, U.R. Associations of B- and C-Raf with Cholesterol, Phosphatidylserine, and Lipid Second Messengers: Preferential Binding of Raf to Artificial Lipid Rafts. J. Biol. Chem. 2002, 277, 24090–24102.
- Park, E.; Rawson, S.; Schmoker, A.; Kim, B.-W.; Oh, S.; Song, K.; Jeon, H.; Eck, M.J. Cryo-EM Structure of a RAS/RAF Recruitment Complex. Nat. Commun. 2023, 14, 4580.
- Terrell, E.M.; Morrison, D.K. Ras-Mediated Activation of the Raf Family Kinases. Cold Spring Harb. Perspect. Med. 2019, 9, a033746.
- Lavoie, H.; Therrien, M. Regulation of RAF Protein Kinases in ERK Signalling. Nat. Rev. Mol. Cell Biol. 2015, 16, 281–298.
- Matallanas, D.; Birtwistle, M.; Romano, D.; Zebisch, A.; Rauch, J.; von Kriegsheim, A.; Kolch, W. Raf Family Kinases: Old Dogs Have Learned New Tricks. Genes Cancer 2011, 2, 232.
- Wellbrock, C.; Karasarides, M.; Marais, R. The RAF Proteins Take Centre Stage. Nat. Rev. Mol. Cell Biol. 2004, 5, 875–885.
- Li, S.; Jang, H.; Zhang, J.; Nussinov, R. Raf-1 Cysteine-Rich Domain Increases the Affinity of K-Ras/Raf at the Membrane, Promoting MAPK Signaling. Structure 2018, 26, 513–525.e2.
- Nguyen, K.; López, C.A.; Neale, C.; Van, Q.N.; Carpenter, T.S.; Di Natale, F.; Travers, T.; Tran, T.H.; Chan, A.H.; Bhatia, H.; et al. Exploring CRD Mobility during RAS/RAF Engagement at the Membrane. Biophys. J. 2022, 121, 3630–3650.
- Fang, Z.; Lee, K.Y.; Huo, K.G.; Gasmi-Seabrook, G.; Zheng, L.; Moghal, N.; Tsao, M.S.; Ikura, M.; Marshall, C.B. Multivalent Assembly of KRAS with the RAS-Binding and Cysteine-Rich Domains of CRAF on the Membrane. Proc. Natl. Acad. Sci. USA 2020, 117, 12101–12108.
- Tran, T.H.; Chan, A.H.; Young, L.C.; Bindu, L.; Neale, C.; Messing, S.; Dharmaiah, S.; Taylor, T.; Denson, J.P.; Esposito, D.; et al. KRAS Interaction with RAF1 RAS-Binding Domain and Cysteine-Rich Domain Provides Insights into RAS-Mediated RAF Activation. Nat. Commun. 2021, 12, 1176.
- Yi, Q.; Peng, J.; Xu, Z.; Liang, Q.; Cai, Y.; Peng, B.; He, Q.; Yan, Y. Spectrum of BRAF Aberrations and Its Potential Clinical Implications: Insights From Integrative Pan-Cancer Analysis. Front. Bioeng. Biotechnol. 2022, 10, 806851.
- Zhao, J.; Luo, Z. Discovery of Raf Family Is a Milestone in Deciphering the Ras-Mediated Intracellular Signaling Pathway. Int. J. Mol. Sci. 2022, 23, 5158.
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF Gene in Human Cancer. Nature 2002, 417, 949–954.
- Ullah, R.; Yin, Q.; Snell, A.H.; Wan, L. RAF-MEK-ERK Pathway in Cancer Evolution and Treatment. Semin. Cancer Biol. 2022, 85, 123–154.
- Wan, P.T.C.; Garnett, M.J.; Roe, S.M.; Lee, S.; Niculescu-Duvaz, D.; Good, V.M.; Project, C.G.; Jones, C.M.; Marshall, C.J.; Springer, C.J.; et al. Mechanism of Activation of the RAF-ERK Signaling Pathway by Oncogenic Mutations of B-RAF. Cell 2004, 116, 855–867.
- Garnett, M.J.; Rana, S.; Paterson, H.; Barford, D.; Marais, R. Wild-Type and Mutant B-RAF Activate C-RAF through Distinct Mechanisms Involving Heterodimerization. Mol. Cell 2005, 20, 963–969.
- Owsley, J.; Stein, M.K.; Porter, J.; In, G.K.; Salem, M.; O’Day, S.; Elliott, A.; Poorman, K.; Gibney, G.; VanderWalde, A. Prevalence of Class I–III BRAF Mutations among 114,662 Cancer Patients in a Large Genomic Database. Exp. Biol. Med. 2021, 246, 31–39.
- Seger, R.; Ahn, N.G.; Posada, J.; Munar, E.S.; Jensen, A.M.; Cooper, J.A.; Cobb, M.H.; Krebs, E.G. Purification and Characterization of Mitogen-Activated Protein Kinase Activator(s) from Epidermal Growth Factor-Stimulated A431 Cells. J. Biol. Chem. 1992, 267, 14373–14381.
- Crews, C.M.; Alessandrini, A.; Erikson, R.L. The Primary Structure of MEK, a Protein Kinase That Phosphorylates the ERK Gene Product. Science 1992, 258, 478–480.
- Kocieniewski, P.; Lipniacki, T. MEK1 and MEK2 Differentially Control the Duration and Amplitude of the ERK Cascade Response. Phys. Biol. 2013, 10, 035006.
- Fukuda, M.; Gotoh, Y.; Nishida, E. Interaction of MAP Kinase with MAP Kinase Kinase: Its Possible Role in the Control of Nucleocytoplasmic Transport of MAP Kinase. EMBO J. 1997, 16, 1901–1908.
- Adachi, M.; Fukuda, M.; Nishida, E. Nuclear Export of MAP Kinase (ERK) Involves a MAP Kinase Kinase (MEK)-Dependent Active Transport Mechanism. J. Cell Biol. 2000, 148, 849–856.
- Zheng, C.F.; Guan, K.L. Activation of MEK Family Kinases Requires Phosphorylation of Two Conserved Ser/Thr Residues. EMBO J. 1994, 13, 1123–1131.
- Nebreda, A.R.; Hill, C.; Gomez, N.; Cohen, P.; Hunt, T. The Protein Kinase Mos Activates MAP Kinase Kinase in Vitro and Stimulates the MAP Kinase Pathway in Mammalian Somatic Cells in Vivo. FEBS Lett. 1993, 333, 183–187.
- Xu, S.; Robbins, D.; Frost, J.; Dang, A.; Lange-Carter, C.; Cobb, M.H. MEKK1 Phosphorylates MEK1 and MEK2 but Does Not Cause Activation of Mitogen-Activated Protein Kinase. Proc. Natl. Acad. Sci. USA 1995, 92, 6808–6812.
- Waterfield, M.R.; Zhang, M.; Norman, L.P.; Sun, S.C. NF-ΚB1/P105 Regulates Lipopolysaccharide-Stimulated MAP Kinase Signaling by Governing the Stability and Function of the Tpl2 Kinase. Mol. Cell 2003, 11, 685–694.
- Jin, L.; Chun, J.; Pan, C.; Li, D.; Lin, R.; Alesi, G.N.; Wang, X.; Kang, H.B.; Song, L.; Wang, D.; et al. MAST1 Drives Cisplatin Resistance in Human Cancers by Rewiring CRaf-Independent MEK Activation. Cancer Cell 2018, 34, 315–330.
- Brunet, A.; Pagès, G.; Pouysségur, J. Growth Factor-Stimulated MAP Kinase Induces Rapid Retrophosphorylation and Inhibition of MAP Kinase Kinase (MEK1). FEBS Lett. 1994, 346, 299–303.
- Eblen, S.T.; Slack-Davis, J.K.; Tarcsafalvi, A.; Parsons, J.T.; Weber, M.J.; Catling, A.D. Mitogen-Activated Protein Kinase Feedback Phosphorylation Regulates MEK1 Complex Formation and Activation during Cellular Adhesion. Mol. Cell. Biol. 2004, 24, 2308–2317.
- Catalanotti, F.; Reyes, G.; Jesenberger, V.; Galabova-Kovacs, G.; De Matos Simoes, R.; Carugo, O.; Baccarini, M. A Mek1-Mek2 Heterodimer Determines the Strength and Duration of the Erk Signal. Nat. Struct. Mol. Biol. 2009, 16, 294–303.
- Bélanger, L.-F.; Roy, S.; Tremblay, M.; Brott, B.; Steff, A.-M.; Mourad, W.; Hugo, P.; Erikson, R.; Charron, J. Mek2 Is Dispensable for Mouse Growth and Development. Mol. Cell. Biol. 2003, 23, 4778–4787.
- Bissonauth, V.; Roy, S.; Gravel, M.; Guillemette, S.; Charron, J. Requirement for Map2k1 (Mek1) in Extra-Embryonic Ectoderm during Placentogenesis. Development 2006, 133, 3429–3440.
- Aoidi, R.; Maltais, A.; Charron, J. Functional Redundancy of the Kinases MEK1 and MEK2: Rescue of the Mek1 Mutant Phenotype by Mek2 Knock-in Reveals a Protein Threshold Effect. Sci. Signal. 2016, 9, ra9.
- Roskoski, R. Invited Review ERK1/2 MAP Kinases: Structure, Function, and Regulation. Pharmacol. Res. 2012, 66, 105–143.
- Eblen, S.T.; Catling, A.D.; Assanah, M.C.; Weber, M.J. Biochemical and Biological Functions of the N-Terminal, Noncatalytic Domain of Extracellular Signal-Regulated Kinase 2. Mol. Cell. Biol. 2001, 21, 249–259.
- Robinson, F.L.; Whitehurst, A.W.; Raman, M.; Cobb, M.H. Identification of Novel Point Mutations in ERK2 That Selectively Disrupt Binding to MEK1. J. Biol. Chem. 2002, 277, 14844–14852.
- Zhou, T.; Sun, L.; Humphreys, J.; Goldsmith, E.J. Docking Interactions Induce Exposure of Activation Loop in the MAP Kinase ERK2. Structure 2006, 14, 1011–1019.
- Sours, K.M.; Xiao, Y.; Ahn, N.G. Extracellular-Regulated Kinase 2 Is Activated by the Enhancement of Hinge Flexibility. J. Mol. Biol. 2014, 426, 1925–1935.
- Lopez, E.D.; Burastero, O.; Arcon, J.P.; Defelipe, L.A.; Ahn, N.G.; Marti, M.A.; Turjanski, A.G. Kinase Activation by Small Conformational Changes. J. Chem. Inf. Model. 2020, 60, 821–832.
- Iverson, D.B.; Xiao, Y.; Jones, D.N.; Eisenmesser, E.Z.; Ahn, N.G. Activation Loop Dynamics Are Coupled to Core Motions in Extracellular Signal-Regulated Kinase-2. Biochemistry 2020, 59, 2698–2706.
- Payne, D.M.; Rossomando, A.J.; Martino, P.; Erickson, A.K.; Her, J.H.; Shabanowitz, J.; Hunt, D.F.; Weber, M.J.; Sturgill, T.W. Identification of the Regulatory Phosphorylation Sites in Pp42/Mitogen-Activated Protein Kinase (MAP Kinase). EMBO J. 1991, 10, 885.
- Robbins, D.J.; Cobb, M.H. Extracellular Signal-Regulated Kinases 2 Autophosphorylates on a Subset of Peptides Phosphorylated in Intact Cells in Response to Insulin and Nerve Growth Factor: Analysis by Peptide Mapping. Mol. Biol. Cell 1992, 3, 299–308.
- Canagarajah, B.J.; Khokhlatchev, A.; Cobb, M.H.; Goldsmith, E.J. Activation Mechanism of the MAP Kinase ERK2 by Dual Phosphorylation. Cell 1997, 90, 859–869.
- Prowse, C.N.; Lew, J. Mechanism of Activation of ERK2 by Dual Phosphorylation. J. Biol. Chem. 2001, 276, 99–103.
- Frémin, C.; Saba-El-Leil, M.K.; Lévesque, K.; Ang, S.L.; Meloche, S. Functional Redundancy of ERK1 and ERK2 MAP Kinases during Development. Cell Rep. 2015, 12, 913–921.
- Saba-El-Leil, M.K.; Frémin, C.; Meloche, S. Redundancy in the World of MAP Kinases: All for One. Front. Cell Dev. Biol. 2016, 4, 67.
- Buscà, R.; Christen, R.; Lovern, M.; Clifford, A.M.; Yue, J.X.; Goss, G.G.; Pouysségur, J.; Lenormand, P. ERK1 and ERK2 Present Functional Redundancy in Tetrapods despite Higher Evolution Rate of ERK1. BMC Evol. Biol. 2015, 15, 179.
- Buscà, R.; Pouysségur, J.; Lenormand, P. ERK1 and ERK2 Map Kinases: Specific Roles or Functional Redundancy? Front. Cell Dev. Biol. 2016, 4, 53.
- Vithayathil, J.; Pucilowska, J.; Goodnough, L.H.; Atit, R.P.; Landreth, G.E. Dentate Gyrus Development Requires ERK Activity to Maintain Progenitor Population and MAPK Pathway Feedback Regulation. J. Neurosci. 2015, 35, 6836–6848.
- Pagès, G.; Guérin, S.; Grall, D.; Bonino, F.; Smith, A.; Anjuere, F.; Auberger, P.; Pouysségur, J. Defective Thymocyte Maturation in P44 MAP Kinase (Erk 1) Knockout Mice. Science 1999, 286, 1374–1377.
- Hatano, N.; Mori, Y.; Oh-hora, M.; Kosugi, A.; Fujikawa, T.; Nakai, N.; Niwa, H.; Miyazaki, J.I.; Hamaoka, T.; Ogata, M. Essential Role for ERK2 Mitogen-Activated Protein Kinase in Placental Development. Genes Cells 2003, 8, 847–856.
- Ünal, E.B.; Uhlitz, F.; Blüthgen, N. A Compendium of ERK Targets. FEBS Lett. 2017, 591, 2607–2615.
- Yang, L.; Zheng, L.; Chng, W.J.; Ding, J.L. Comprehensive Analysis of ERK1/2 Substrates for Potential Combination Immunotherapies. Trends Pharmacol. Sci. 2019, 40, 897–910.
- Johnson, J.L.; Yaron, T.M.; Huntsman, E.M.; Kerelsky, A.; Song, J.; Regev, A.; Lin, T.Y.; Liberatore, K.; Cizin, D.M.; Cohen, B.M.; et al. An Atlas of Substrate Specificities for the Human Serine/Threonine Kinome. Nature 2023, 613, 759–766.
- Santini, C.C.; Longden, J.; Schoof, E.M.; Simpson, C.D.; Jeschke, G.R.; Creixell, P.; Kim, J.; Wu, X.; Turk, B.E.; Rosen, N.; et al. Global View of the RAF-MEK-ERK Module and Its Immediate Downstream Effectors. Sci. Rep. 2019, 9, 10865.
- Tanoue, T.; Maeda, R.; Adachi, M.; Nishida, E. Identification of a Docking Groove on ERK and P38 MAP Kinases That Regulates the Specificity of Docking Interactions. EMBO J. 2001, 20, 466–479.
- Tanoue, T.; Adachi, M.; Moriguchi, T.; Nishida, E. A Conserved Docking Motif in MAP Kinases Common to Substrates, Activators and Regulators. Nat. Cell Biol. 2000, 2, 110–116.
- Jacobs, D.; Glossip, D.; Xing, H.; Muslin, A.J.; Kornfeld, K. Multiple Docking Sites on Substrate Proteins Form a Modular System That Mediates Recognition by ERK MAP Kinase. Genes Dev. 1999, 13, 163–175.
- Ahn, N.G. PORE-Ing over ERK Substrates. Nat. Struct. Mol. Biol. 2009, 16, 1004–1005.
- Lee, T.; Hoofnagle, A.N.; Kabuyama, Y.; Stroud, J.; Min, X.; Goldsmith, E.J.; Chen, L.; Resing, K.A.; Ahn, N.G. Docking Motif Interactions in Map Kinases Revealed by Hydrogen Exchange Mass Spectrometry. Mol. Cell 2004, 14, 43–55.
- Piserchio, A.; Ramakrishan, V.; Wang, H.; Kaoud, T.S.; Arshava, B.; Dutta, K.; Dalby, K.N.; Ghose, R. Structural and Dynamic Features of F-Recruitment Site Driven Substrate Phosphorylation by ERK2. Sci. Rep. 2015, 5, 11127.
- Casar, B.; Crespo, P. ERK Signals: Scaffolding Scaffolds? Front. Cell Dev. Biol. 2016, 4, 49.
- Matsubayashi, Y.; Fukuda, M.; Nishida, E. Evidence for Existence of a Nuclear Pore Complex-Mediated, Cytosol-Independent Pathway of Nuclear Translocation of ERK MAP Kinase in Permeabilized Cells. J. Biol. Chem. 2001, 276, 41755–41760.
- Whitehurst, A.W.; Wilsbacher, J.L.; You, Y.; Luby-Phelps, K.; Moore, M.S.; Cobb, M.H. ERK2 Enters the Nucleus by a Carrier-Independent Mechanism. Proc. Natl. Acad. Sci. USA 2002, 99, 7496–7501.
- Vomastek, T.; Iwanicki, M.P.; Burack, W.R.; Tiwari, D.; Kumar, D.; Parsons, J.T.; Weber, M.J.; Nandicoori, V.K. Extracellular Signal-Regulated Kinase 2 (ERK2) Phosphorylation Sites and Docking Domain on the Nuclear Pore Complex Protein Tpr Cooperatively Regulate ERK2-Tpr Interaction. Mol. Cell. Biol. 2008, 28, 6954–6966.
- Atias, K.S.; Soudah, N.; Engelberg, D. Mutations That Confer Drug-Resistance, Oncogenicity and Intrinsic Activity on the ERK MAP Kinases—Current State of the Art. Cells 2020, 9, 129.
- Emrick, M.A.; Hoofnagle, A.N.; Miller, A.S.; Ten Eyck, L.F.; Ahn, N.G. Constitutive Activation of Extracellular Signal-Regulated Kinase 2 by Synergistic Point Mutations. J. Biol. Chem. 2001, 276, 46469–46479.
- Levin-Salomon, V.; Kogan, K.; Ahn, N.G.; Livnah, O.; Engelberg, D. Isolation of Intrinsically Active (MEK-Independent) Variants of the ERK Family of Mitogen-Activated Protein (MAP) Kinases. J. Biol. Chem. 2008, 283, 34500–34510.
- Smorodinsky-Atias, K.; Goshen-Lago, T.; Goldberg-Carp, A.; Melamed, D.; Shir, A.; Mooshayef, N.; Beenstock, J.; Karamansha, Y.; Darlyuk-Saadon, I.; Livnah, O.; et al. Intrinsically Active Variants of Erk Oncogenically Transform Cells and Disclose Unexpected Autophosphorylation Capability That Is Independent of TEY Phosphorylation. Mol. Biol. Cell 2015, 27, 1026–1039.
- Kushnir, T.; Bar-Cohen, S.; Mooshayef, N.; Lange, R.; Bar-Sinai, A.; Rozen, H.; Salzberg, A.; Engelberg, D.; Paroush, Z. An Activating Mutation in ERK Causes Hyperplastic Tumors in a Scribble Mutant Tissue in Drosophila. Genetics 2020, 214, 109–120.
- COSMIC: Catalog of Somatic Mutations in Cancer. Available online: https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=MAPK3 (accessed on 29 September 2023).
- COSMIC: Catalog of Somatic Mutations in Cancer. Available online: https://cancer.sanger.ac.uk/cosmic/gene/analysis?ln=MAPK1_ENST00000398822 (accessed on 29 September 2023).
- Arvind, R.; Shimamoto, H.; Momose, F.; Amagasa, T.; Omura, K.; Tsuchida, N. A Mutation in the Common Docking Domain of ERK2 in a Human Cancer Cell Line, Which Was Associated with Its Constitutive Phosphorylation. Int. J. Oncol. 2005, 27, 1499–1504.
- Lavoie, H.; Gagnon, J.; Therrien, M. ERK Signalling: A Master Regulator of Cell Behaviour, Life and Fate. Nat. Rev. Mol. Cell Biol. 2020, 21, 607–632.
- Fey, D.; Croucher, D.R.; Kolch, W.; Kholodenko, B.N. Crosstalk and Signaling Switches in Mitogen-Activated Protein Kinase Cascades. Front. Physiol. 2012, 3, 355.
- Saba-El-Leil, M.K.; Vella, F.D.J.; Vernay, B.; Voisin, L.; Chen, L.; Labrecque, N.; Ang, S.L.; Meloche, S. An Essential Function of the Mitogen-Activated Protein Kinase Erk2 in Mouse Trophoblast Development. EMBO Rep. 2003, 4, 964–968.
- Pucilowska, J.; Puzerey, P.A.; Karlo, J.C.; Galán, R.F.; Landreth, G.E. Development/Plasticity/Repair Disrupted ERK Signaling during Cortical Development Leads to Abnormal Progenitor Proliferation, Neuronal and Network Excitability and Behavior, Modeling Human Neuro-Cardio-Facial-Cutaneous and Related Syndromes. J. Neurosci. 2012, 32, 8663–8677.
- Seaberg, B.; Henslee, G.; Wang, S.; Paez-Colasante, X.; Landreth, G.E.; Rimer, M. Muscle-Derived Extracellular Signal-Regulated Kinases 1 and 2 Are Required for the Maintenance of Adult Myofibers and Their Neuromuscular Junctions. Mol. Cell. Biol. 2015, 35, 1238–1253.
- Kosako, H.; Yamaguchi, N.; Aranami, C.; Ushiyama, M.; Kose, S.; Imamoto, N.; Taniguchi, H.; Nishida, E.; Hattori, S. Phosphoproteomics Reveals New ERK MAP Kinase Targets and Links ERK to Nucleoporin-Mediated Nuclear Transport. Nat. Struct. Mol. Biol. 2009, 16, 1026–1035.
- Courcelles, M.; Frémin, C.; Voisin, L.; Lemieux, S.; Meloche, S.; Thibault, P. Phosphoproteome Dynamics Reveal Novel ERK1/2 MAP Kinase Substrates with Broad Spectrum of Functions. Mol. Syst. Biol. 2013, 9, 669.
- Carlson, S.M.; Chouinard, C.R.; Labadorf, A.; Lam, C.J.; Schmelzle, K.; Fraenkel, E.; White, F.M. Large-Scale Discovery of ERK2 Substrates Identifies ERK-Mediated Transcriptional Regulation by ETV3. Sci. Signal. 2011, 4, rs11.
- Zeke, A.; Bastys, T.; Alexa, A.; Garai, Á.; Mészáros, B.; Kirsch, K.; Dosztányi, Z.; Kalinina, O.V.; Reményi, A. Systematic Discovery of Linear Binding Motifs Targeting an Ancient Protein Interaction Surface on MAP Kinases. Mol. Syst. Biol. 2015, 11, 837.
- Gógl, G.; Biri-Kovács, B.; Póti, Á.L.; Vadászi, H.; Szeder, B.; Bodor, A.; Schlosser, G.; Ács, A.; Turiák, L.; Buday, L.; et al. Dynamic Control of RSK Complexes by Phosphoswitch-Based Regulation. FEBS J. 2018, 285, 46–71.
- Lawrence, M.C.; McGlynn, K.; Park, B.H.; Cobb, M.H. ERK1/2-Dependent Activation of Transcription Factors Required for Acute and Chronic Effects of Glucose on the Insulin Gene Promoter. J. Biol. Chem. 2005, 280, 26751–26759.
- Lawrence, M.; Shao, C.; Duan, L.; McGlynn, K.; Cobb, M.H. The Protein Kinases ERK1/2 and Their Roles in Pancreatic Beta Cells. Acta Physiol. 2008, 192, 11–17.
- Chamberlain, C.E.; Scheel, D.W.; McGlynn, K.; Kim, H.; Miyatsuka, T.; Wang, J.; Nguyen, V.; Zhao, S.; Mavropoulos, A.; Abraham, A.G.; et al. Menin Determines K-RAS Proliferative Outputs in Endocrine Cells. J. Clin. Investig. 2014, 124, 4093–4101.
- Konieczkowski, D.J.; Johannessen, C.M.; Abudayyeh, O.; Kim, J.W.; Cooper, Z.A.; Piris, A.; Frederick, D.T.; Barzily-Rokni, M.; Straussman, R.; Haq, R.; et al. A Melanoma Cell State Distinction Influences Sensitivity to MAPK Pathway Inhibitors. Cancer Discov. 2014, 4, 816–827.
- Wang, Y.; Yang, F.; Fu, Y.; Huang, X.; Wang, W.; Jiang, X.; Gritsenko, M.A.; Zhao, R.; Monore, M.E.; Pertz, O.C.; et al. Spatial Phosphoprotein Profiling Reveals a Compartmentalized Extracellular Signal-Regulated Kinase Switch Governing Neurite Growth and Retraction. J. Biol. Chem. 2011, 286, 18190–18201.
- Drosten, M.; Dhawahir, A.; Sum, E.Y.M.; Urosevic, J.; Lechuga, C.G.; Esteban, L.M.; Castellano, E.; Guerra, C.; Santos, E.; Barbacid, M. Genetic Analysis of Ras Signalling Pathways in Cell Proliferation, Migration and Survival. EMBO J. 2010, 29, 1091–1104.
- Wang, Z. Regulation of Cell Cycle Progression by Growth Factor-Induced Cell Signaling. Cells 2021, 10, 3327.
- Ginzberg, M.B.; Kafri, R.; Kirschner, M. On Being the Right (Cell) Size. Science 2015, 348, 1245075.
- Chambard, J.C.; Lefloch, R.; Pouysségur, J.; Lenormand, P. ERK Implication in Cell Cycle Regulation. Biochim. Biophys. Acta-Mol. Cell Res. 2007, 1773, 1299–1310.
- Kotsis, D.H.; Masko, E.M.; Sigoillot, F.D.; Gregorio, R.D.; Guy-Evans, H.I.; Evans, D.R. Protein Kinase A Phosphorylation of the Multifunctional Protein CAD Antagonizes Activation by the MAP Kinase Cascade. Mol. Cell. Biochem. 2007, 301, 69–81.
- Stefanovsky, V.; Langlois, F.; Gagnon-Kugler, T.; Rothblum, L.I.; Moss, T. Growth Factor Signaling Regulates Elongation of RNA Polymerase I Transcription in Mammals via UBF Phosphorylation and R-Chromatin Remodeling. Mol. Cell 2006, 21, 629–639.
- Felton-Edkins, Z.A.; Fairley, J.A.; Graham, E.L.; Johnston, I.M.; White, R.J.; Scott, P.H. The Mitogen-Activated Protein (MAP) Kinase ERK Induces TRNA Synthesis by Phosphorylating TFIIIB. EMBO J. 2003, 22, 2422–2432.
- Scheper, G.C.; Morrice, N.A.; Kleijn, M.; Proud, C.G. The Mitogen-Activated Protein Kinase Signal-Integrating Kinase Mnk2 Is a Eukaryotic Initiation Factor 4E Kinase with High Levels of Basal Activity in Mammalian Cells. Mol. Cell. Biol. 2001, 21, 743–754.
- Kalous, J.; Tetkova, A.; Kubelka, M.; Susor, A. Importance of ERK1/2 in Regulation of Protein Translation during Oocyte Meiosis. Int. J. Mol. Sci. 2018, 19, 698.
- Jiang, J.C.; Zhang, H.; Cao, L.R.; Dai, X.X.; Zhao, L.W.; Liu, H.B.; Fan, H.Y. Oocyte Meiosis-Coupled Poly(A) Polymerase α Phosphorylation and Activation Trigger Maternal MRNA Translation in Mice. Nucleic Acids Res. 2021, 49, 5867.
- Khoo, S.; Griffen, S.C.; Xia, Y.; Baer, R.J.; German, M.S.; Cobb, M.H. Regulation of Insulin Gene Transcription by ERK1 and ERK2 in Pancreatic β Cells. J. Biol. Chem. 2003, 278, 32969–32977.
- Arnette, D.; Gibson, T.B.; Lawrence, M.C.; January, B.; Khoo, S.; McGlynn, K.; Vanderbilt, C.A.; Cobb, M.H. Regulation of ERK1 and ERK2 by Glucose and Peptide Hormones in Pancreatic β Cells. J. Biol. Chem. 2003, 278, 32517–32525.
- Ritt, D.A.; Monson, D.M.; Specht, S.I.; Morrison, D.K. Impact of Feedback Phosphorylation and Raf Heterodimerization on Normal and Mutant B-Raf Signaling. Mol. Cell. Biol. 2010, 30, 806–819.
- Duan, L.; Cobb, M.H. Calcineurin Increases Glucose Activation of ERK1/2 by Reversing Negative Feedback. Proc. Natl. Acad. Sci. USA 2010, 107, 22314–22319.
- Nichols, J.; Smith, A. Naive and Primed Pluripotent States. Cell Stem Cell 2009, 4, 487–492.
- Huang, Y.; Osorno, R.; Tsakiridis, A.; Wilson, V. In Vivo Differentiation Potential of Epiblast Stem Cells Revealed by Chimeric Embryo Formation. Cell Rep. 2012, 2, 1571–1578.
- Williams, R.L.; Hilton, D.J.; Pease, S.; Willson, T.A.; Stewart, C.L.; Gearing, D.P.; Wagner, E.F.; Metcalf, D.; Nicola, N.A.; Gough, N.M. Myeloid Leukaemia Inhibitory Factor Maintains the Developmental Potential of Embryonic Stem Cells. Nature 1988, 336, 684–687.
- Niwa, H.; Ogawa, K.; Shimosato, D.; Adachi, K. A Parallel Circuit of LIF Signalling Pathways Maintains Pluripotency of Mouse ES Cells. Nature 2009, 460, 118–122.
- Ying, Q.L.; Nichols, J.; Chambers, I.; Smith, A. BMP Induction of Id Proteins Suppresses Differentiation and Sustains Embryonic Stem Cell Self-Renewal in Collaboration with STAT3. Cell 2003, 115, 281–292.
- Chazaud, C.; Yamanaka, Y.; Pawson, T.; Rossant, J. Early Lineage Segregation between Epiblast and Primitive Endoderm in Mouse Blastocysts through the Grb2-MAPK Pathway. Dev. Cell 2006, 10, 615–624.
- Qi, X.; Li, T.G.; Hao, J.; Hu, J.; Wang, J.; Simmons, H.; Miura, S.; Mishina, Y.; Zhao, G.Q. BMP4 Supports Self-Renewal of Embryonic Stem Cells by Inhibiting Mitogen-Activated Protein Kinase Pathways. Proc. Natl. Acad. Sci. USA 2004, 101, 6027–6032.
- Li, Z.; Fei, T.; Zhang, J.; Zhu, G.; Wang, L.; Lu, D.; Chi, X.; Teng, Y.; Hou, N.; Yang, X.; et al. BMP4 Signaling Acts via Dual-Specificity Phosphatase 9 to Control ERK Activity in Mouse Embryonic Stem Cells. Cell Stem Cell 2012, 10, 171–182.
- Sato, N.; Meijer, L.; Skaltsounis, L.; Greengard, P.; Brivanlou, A.H. Maintenance of Pluripotency in Human and Mouse Embryonic Stem Cells through Activation of Wnt Signaling by a Pharmacological GSK-3-Specific Inhibitor. Nat. Med. 2004, 10, 55–63.
- Bone, H.K.; Damiano, T.; Bartlett, S.; Perry, A.; Letchford, J.; Ripoll, Y.S.; Nelson, A.S.; Welham, M.J. Involvement of GSK-3 in Regulation of Murine Embryonic Stem Cell Self-Renewal Revealed by a Series of Bisindolylmaleimides. Chem. Biol. 2009, 16, 15–27.
- Ying, Q.L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The Ground State of Embryonic Stem Cell Self-Renewal. Nature 2008, 453, 519–523.
- Silva, J.; Barrandon, O.; Nichols, J.; Kawaguchi, J.; Theunissen, T.W.; Smith, A. Promotion of Reprogramming to Ground State Pluripotency by Signal Inhibition. PLoS Biol. 2008, 6, 2237–2247.
- Stavridis, M.P.; Simon Lunn, J.; Collins, B.J.; Storey, K.G. A Discrete Period of FGF-Induced Erk1/2 Signalling Is Required for Vertebrate Neural Specification. Development 2007, 134, 2889–2894.
- Hamilton, W.B.; Kaji, K.; Kunath, T. ERK2 Suppresses Self-Renewal Capacity of Embryonic Stem Cells, but Is Not Required for Multi-Lineage Commitment. PLoS ONE 2013, 8, e60907.
- Martin, G.R. Isolation of a Pluripotent Cell Line from Early Mouse Embryos Cultured in Medium Conditioned by Teratocarcinoma Stem Cells. Proc. Natl. Acad. Sci. USA 1981, 78, 7634–7638.
- Evans, M.J.; Kaufman, M.H. Establishment in Culture of Pluripotential Cells from Mouse Embryos. Nature 1981, 292, 154–156.
- Brons, I.G.M.; Smithers, L.E.; Trotter, M.W.B.; Rugg-Gunn, P.; Sun, B.; Chuva De Sousa Lopes, S.M.; Howlett, S.K.; Clarkson, A.; Ahrlund-Richter, L.; Pedersen, R.A.; et al. Derivation of Pluripotent Epiblast Stem Cells from Mammalian Embryos. Nature 2007, 448, 191–195.
- Vallier, L.; Touboul, T.; Chng, Z.; Brimpari, M.; Hannan, N.; Millan, E.; Smithers, L.E.; Trotter, M.; Rugg-Gunn, P.; Weber, A.; et al. Early Cell Fate Decisions of Human Embryonic Stem Cells and Mouse Epiblast Stem Cells Are Controlled by the Same Signalling Pathways. PLoS ONE 2009, 4, e6082.
- Weinberger, L.; Ayyash, M.; Novershtern, N.; Hanna, J.H. Dynamic Stem Cell States: Naive to Primed Pluripotency in Rodents and Humans. Nat. Rev. Mol. Cell Biol. 2016, 17, 155–169.
- Tesar, P.J.; Chenoweth, J.G.; Brook, F.A.; Davies, T.J.; Evans, E.P.; Mack, D.L.; Gardner, R.L.; McKay, R.D.G. New Cell Lines from Mouse Epiblast Share Defining Features with Human Embryonic Stem Cells. Nature 2007, 448, 196–199.
- Hu, S.; Xie, Z.; Onishi, A.; Yu, X.; Jiang, L.; Lin, J.; Rho, H.S.; Woodard, C.; Wang, H.; Jeong, J.S.; et al. Profiling the Human Protein-DNA Interactome Reveals ERK2 as a Transcriptional Repressor of Interferon Signaling. Cell 2009, 139, 610–622.
- Tripathi, P.; Sahoo, N.; Ullah, U.; Kallionpää, H.; Suneja, A.; Lahesmaa, R.; Rao, K.V.S. A Novel Mechanism for ERK-Dependent Regulation of IL4 Transcription during Human Th2-Cell Differentiation. Immunol. Cell Biol. 2012, 90, 676–687.
- Agudo-Ibáñez, L.; Morante, M.; García-Gutiérrez, L.; Quintanilla, A.; Rodríguez, J.; Muñoz, A.; León, J.; Crespo, P. ERK2 Stimulates MYC Transcription by Anchoring CDK9 to the MYC Promoter in a Kinase Activity-Independent Manner. Sci. Signal. 2023, 16, eadg4193.
- Fjeld, C.C.; Rice, A.E.; Kim, Y.; Gee, K.R.; Denu, J.M. Mechanistic Basis for Catalytic Activation of Mitogen-Activated Protein Kinase Phosphatase 3 by Extracellular Signal-Regulated Kinase. J. Biol. Chem. 2000, 275, 6749–6757.
- Camps, M.; Nichols, A.; Gillieron, C.; Antonsson, B.; Muda, M.; Chabert, C.; Boschert, U.; Arkinstall, S. Catalytic Activation of the Phosphatase MKP-3 by ERK2 Mitogen-Activated Protein Kinase. Science 1998, 280, 1262–1265.
- Lake, D.; Corrêa, S.A.L.; Müller, J. Negative Feedback Regulation of the ERK1/2 MAPK Pathway. Cell. Mol. Life Sci. 2016, 73, 4397–4413.
- Dougherty, M.K.; Müller, J.; Ritt, D.A.; Zhou, M.; Zhou, X.Z.; Copeland, T.D.; Conrads, T.P.; Veenstra, T.D.; Lu, K.P.; Morrison, D.K. Regulation of Raf-1 by Direct Feedback Phosphorylation. Mol. Cell 2005, 17, 215–224.
- Hekman, M.; Fischer, A.; Wennogle, L.P.; Wang, Y.K.; Campbell, S.L.; Rapp, U.R. Novel C-Raf Phosphorylation Sites: Serine 296 and 301 Participate in Raf Regulation. FEBS Lett. 2005, 579, 464–468.
- Brummer, T.; Naegele, H.; Reth, M.; Misawa, Y. Identification of Novel ERK-Mediated Feedback Phosphorylation Sites at the C-Terminus of B-Raf. Oncogene 2003, 22, 8823–8834.
- Rushworth, L.K.; Hindley, A.D.; O’Neill, E.; Kolch, W. Regulation and Role of Raf-1/B-Raf Heterodimerization. Mol. Cell. Biol. 2006, 26, 2262–2272.
- Wilhelm, M.; Schlegl, J.; Hahne, H.; Gholami, A.M.; Lieberenz, M.; Savitski, M.M.; Ziegler, E.; Butzmann, L.; Gessulat, S.; Marx, H.; et al. Mass-Spectrometry-Based Draft of the Human Proteome. Nature 2014, 509, 582–587.
- Fujioka, A.; Terai, K.; Itoh, R.E.; Aoki, K.; Nakamura, T.; Kuroda, S.; Nishida, E.; Matsuda, M. Dynamics of the Ras/ERK MAPK Cascade as Monitored by Fluorescent Probes. J. Biol. Chem. 2006, 281, 8917–8926.
- Choi, K.Y.; Satterberg, B.; Lyons, D.M.; Elion, E.A. Ste5 Tethers Multiple Protein Kinases in the MAP Kinase Cascade Required for Mating in S. Cerevisiae. Cell 1994, 78, 499–512.
- Yablonski, D.; Marbach, I.; Levitzki, A. Dimerization of Ste5, a Mitogen-Activated Protein Kinase Cascade Scaffold Protein, Is Required for Signal Transduction. Proc. Natl. Acad. Sci. USA 1996, 93, 13864–13869.
- Mataraza, J.M.; Briggs, M.W.; Li, Z.; Entwistle, A.; Ridley, A.J.; Sacks, D.B. IQGAP1 Promotes Cell Motility and Invasion. J. Biol. Chem. 2003, 278, 41237–41245.
- Ishibe, S.; Joly, D.; Liu, Z.X.; Cantley, L.G. Paxillin Serves as an ERK-Regulated Scaffold for Coordinating FAK and Rac Activation in Epithelial Morphogenesis. Mol. Cell 2004, 16, 257–267.
- Yin, G.; Zheng, Q.; Yan, C.; Berk, B.C. GIT1 Is a Scaffold for ERK1/2 Activation in Focal Adhesions. J. Biol. Chem. 2005, 280, 27705–27712.
- Witzel, F.; Maddison, L.; Blüthgen, N. How Scaffolds Shape MAPK Signaling: What We Know and Opportunities for Systems Approaches. Front. Physiol. 2012, 3, 475.
- Scott, A.; Haystead, C.M.; Haystead, T.A. Purification of a 12,020-Dalton Protein That Enhances the Activation of Mitogen-Activated Protein (MAP) Kinase by MAP Kinase Kinase. J. Biol. Chem. 1995, 270, 24540–24547.
- Levchenko, A.; Bruck, J.; Sternberg, P.W. Scaffold Proteins May Biphasically Affect the Levels of Mitogen-Activated Protein Kinase Signaling and Reduce Its Threshold Properties. Proc. Natl. Acad. Sci. USA 2000, 97, 5818–5823.
- Ha, S.H.; Kim, S.Y.; Ferrell, J.E. The Prozone Effect Accounts for the Paradoxical Function of the Cdk-Binding Protein Suc1/Cks. Cell Rep. 2016, 14, 1408–1421.
- Locasale, J.W.; Shaw, A.S.; Chakraborty, A.K. Scaffold Proteins Confer Diverse Regulatory Properties to Protein Kinase Cascades. Proc. Natl. Acad. Sci. USA 2007, 104, 13307–13312.
- Heinrich, R.; Neel, B.G.; Rapoport, T.A. Mathematical Models of Protein Kinase Signal Transduction. Mol. Cell 2002, 9, 957–970.
- Chapman, S.A.; Asthagiri, A.R. Quantitative Effect of Scaffold Abundance on Signal Propagation. Mol. Syst. Biol. 2009, 5, 313.
- Nguyen, A.; Burack, W.R.; Stock, J.L.; Kortum, R.; Chaika, O.V.; Afkarian, M.; Muller, W.J.; Murphy, K.M.; Morrison, D.K.; Lewis, R.E.; et al. Kinase Suppressor of Ras (KSR) Is a Scaffold Which Facilitates Mitogen-Activated Protein Kinase Activation in Vivo. Mol. Cell. Biol. 2002, 22, 3035–3045.
- Lozano, J.; Xing, R.; Cai, Z.; Jensen, H.L.; Trempus, C.; Mark, W.; Cannon, R.; Kolesnick, R. Deficiency of Kinase Suppressor of Ras1 Prevents Oncogenic Ras Signaling in Mice. Cancer Res. 2003, 63, 4232–4238.
- Roy, M.; Li, Z.; Sacks, D.B. IQGAP1 Binds ERK2 and Modulates Its Activity. J. Biol. Chem. 2004, 279, 17329–17337.
- Jameson, K.L.; Mazur, P.K.; Zehnder, A.M.; Zhang, J.; Zarnegar, B.; Sage, J.; Khavari, P.A. IQGAP1 Scaffold-Kinase Interaction Blockade Selectively Targets RAS-MAP Kinase-Driven Tumors. Nat. Med. 2013, 19, 626–630.
- Sharma, C.; Vomastek, T.; Tarcsafalvi, A.; Catling, A.D.; Schaeffer, H.J.; Eblen, S.T.; Weber, M.J. MEK Partner 1 (MP1): Regulation of Oligomerization in MAP Kinase Signaling. J. Cell. Biochem. 2005, 94, 708–719.
- Teis, D.; Taub, N.; Kurzbauer, R.; Hilber, D.; De Araujo, M.E.; Erlacher, M.; Offterdinger, M.; Villunger, A.; Geley, S.; Bohn, G.; et al. P14-MP1-MEK1 Signaling Regulates Endosomal Traffic and Cellular Proliferation during Tissue Homeostasis. J. Cell Biol. 2006, 175, 861–868.
- Vomastek, T.; Schaeffer, H.J.; Tarcsafalvi, A.; Smolkin, M.E.; Bissonette, E.A.; Weber, M.J. Modular Construction of a Signaling Scaffold: MORG1 Interacts with Components of the ERK Cascade and Links ERK Signaling to Specific Agonists. Proc. Natl. Acad. Sci. USA 2004, 101, 6981–6986.
- Schiefermeier, N.; Scheffler, J.M.; de Araujo, M.E.G.; Stasyk, T.; Yordanov, T.; Ebner, H.L.; Offterdinger, M.; Munck, S.; Hess, M.W.; Wickström, S.A.; et al. The Late Endosomal P14-MP1 (LAMTOR2/3) Complex Regulates Focal Adhesion Dynamics during Cell Migration. J. Cell Biol. 2014, 205, 525–540.
- Ren, Y.; Meng, S.; Mei, L.; Zhao, Z.J.; Jove, R.; Wu, J. Roles of Gab1 and SHP2 in Paxillin Tyrosine Dephosphorylation and Src Activation in Response to Epidermal Growth Factor. J. Biol. Chem. 2004, 279, 8497–8505.
- Feigin, M.E.; Xue, B.; Hammell, M.C.; Muthuswamy, S.K. G-Protein-Coupled Receptor GPR161 Is Overexpressed in Breast Cancer and Is a Promoter of Cell Proliferation and Invasion. Proc. Natl. Acad. Sci. USA 2014, 111, 4191–4196.
- Martín-Vega, A.; Ruiz-Peinado, L.; García-Gómez, R.; Herrero, A.; de la Fuente-Vivas, D.; Parvathaneni, S.; Caloto, R.; Morante, M.; von Kriegsheim, A.; Bustelo, X.R.; et al. Scaffold Coupling: ERK Activation by Trans-Phosphorylation across Different Scaffold Protein Species. Sci. Adv. 2023, 9, eadd7969.
- Therrien, M.; Chang, H.C.; Solomon, N.M.; Karim, F.D.; Wassarman, D.A.; Rubin, G.M. KSR, a Novel Protein Kinase Required for RAS Signal Transduction. Cell 1995, 83, 879–888.
- Kornfeld, K.; Hom, D.B.; Horvitz, H.R. The Ksr-1 Gene Encodes a Novel Protein Kinase Involved in Ras-Mediated Signaling in C. Elegans. Cell 1995, 83, 903–913.
- Sundaram, M.; Han, M. The C. Elegans Ksr-1 Gene Encodes a Novel Raf-Related Kinase Involved in Ras-Mediated Signal Transduction. Cell 1995, 83, 889–901.
- Channavajhala, P.L.; Wu, L.; Cuozzo, J.W.; Hall, J.P.; Liu, W.; Lin, L.L.; Zhang, Y. Identification of a Novel Human Kinase Supporter of Ras (HKSR-2) That Functions as a Negative Regulator of Cot (Tp12) Signaling. J. Biol. Chem. 2003, 278, 47089–47097.
- Costanzo-Garvey, D.L.; Pfluger, P.T.; Dougherty, M.K.; Stock, J.L.; Boehm, M.; Chaika, O.; Fernandez, M.R.; Fisher, K.; Kortum, R.L.; Hong, E.G.; et al. KSR2 Is an Essential Regulator of AMP Kinase, Energy Expenditure, and Insulin Sensitivity. Cell Metab. 2009, 10, 366–378.
- Pilbrow, A.P. Discovery of an Obesity Susceptibility Gene, KSR2, Provides New Insight into Energy Homeostasis Pathways. Circ. Cardiovasc. Genet. 2014, 7, 218–219.
- Weinmaster, G.; Zoller, M.J.; Pawson, T. A Lysine in the ATP-Binding Site of P130gag-Fps Is Essential for Protein-Tyrosine Kinase Activity. EMBO J. 1986, 5, 69–76.
- Hanks, S.K.; Quinn, A.M.; Hunter, T. The Protein Kinase Family: Conserved Features and Deduced Phylogeny of the Catalytic Domains. Science 1988, 241, 42–52.
- Boudeau, J.; Miranda-Saavedra, D.; Barton, G.J.; Alessi, D.R. Emerging Roles of Pseudokinases. Trends Cell Biol. 2006, 16, 443–452.
- Hu, J.; Yu, H.; Kornev, A.P.; Zhao, J.; Filbert, E.L.; Taylor, S.S.; Shaw, A.S. Mutation That Blocks ATP Binding Creates a Pseudokinase Stabilizing the Scaffolding Function of Kinase Suppressor of Ras, CRAF and BRAF. Proc. Natl. Acad. Sci. USA 2011, 108, 6067–6072.
- Eyers, P.A.; Murphy, J.M. Exploring Kinomes: Pseudokinases and beyond: Dawn of the Dead: Protein Pseudokinases Signal New Adventures in Cell Biology. Biochem. Soc. Trans. 2013, 41, 969–974.
- Zhang, H.; Koo, C.Y.; Stebbing, J.; Giamas, G. The Dual Function of KSR1: A Pseudokinase and Beyond. Biochem. Soc. Trans. 2013, 41, 1078–1082.
- Brennan, D.F.; Dar, A.C.; Hertz, N.T.; Chao, W.C.H.; Burlingame, A.L.; Shokat, K.M.; Barford, D. A Raf-Induced Allosteric Transition of KSR Stimulates Phosphorylation of MEK. Nature 2011, 472, 366–369.
- Zhang, Y.; Bei, Y.; Delikat, S.; Bayoumy, S.; Lin, X.H.; Basu, S.; McGinley, M.; Chan-Hui, P.-Y.; Lichenstein, H.; Kolesnick, R. Kinase Suppressor of Ras Is Ceramide-Activated Protein Kinase. Cell 1997, 89, 63–72.
- Goettel, J.A.; Liang, D.; Hilliard, V.C.; Edelblum, K.L.; Broadus, M.R.; Gould, K.L.; Hanks, S.K.; Polk, D.B. KSR1 Is a Functional Protein Kinase Capable of Serine Autophosphorylation and Direct Phosphorylation of MEK1. Exp. Cell Res. 2011, 317, 452–463.
- Michaud, N.R.; Therrien, M.; Cacace, A.; Edsall, L.C.; Spiegel, S.; Rubin, G.M.; Morrison, D.K. KSR Stimulates Raf-1 Activity in a Kinase-Independent Manner. Proc. Natl. Acad. Sci. USA 1997, 94, 12792–12796.
- Stewart, S.; Sundaram, M.; Zhang, Y.; Lee, J.; Han, M.; Guan, K.-L. Kinase Suppressor of Ras Forms a Multiprotein Signaling Complex and Modulates MEK Localization. Mol. Cell. Biol. 1999, 19, 5523–5534.
- Roy, F.; Laberge, G.; Douziech, M.; Ferland-McCollough, D.; Therrien, M. KSR Is a Scaffold Required for Activation of the ERK/MAPK Module. Genes Dev. 2002, 16, 427–438.
- Paniagua, G.; Jacob, H.K.C.; Brehey, O.; García-Alonso, S.; Lechuga, C.G.; Pons, T.; Musteanu, M.; Guerra, C.; Drosten, M.; Barbacid, M. KSR Induces RAS-Independent MAPK Pathway Activation and Modulates the Efficacy of KRAS Inhibitors. Mol. Oncol. 2022, 16, 3066–3081.
- Therrien, M.; Michaud, N.R.; Rubin, G.M.; Morrison, D.K. KSR Modulates Signal Propagation within the MAPK Cascade. Genes Dev. 1996, 10, 2684–2695.
- Denouel-Galy, A.; Douville, E.M.; Warne, P.H.; Papin, C.; Laugier, D.; Calothy, G.; Downward, J.; Eychène, A. Murine Ksr Interacts with MEK and Inhibits Ras-Induced Transformation. Curr. Biol. 1998, 8, 46–55.
- Xing, H.; Kornfeld, K.; Muslin, A.J. The Protein Kinase KSR Interacts with 14-3-3 Protein and Raf. Curr. Biol. 1997, 7, 294–300.
- Yu, W.; Fantl, W.J.; Harrowe, G.; Williams, L.T. Regulation of the MAP Kinase Pathway by Mammalian Ksr through Direct Interaction with MEK and ERK. Curr. Biol. 1998, 8, 56–64.
- Lavoie, H.; Sahmi, M.; Maisonneuve, P.; Marullo, S.A.; Thevakumaran, N.; Jin, T.; Kurinov, I.; Sicheri, F.; Therrien, M. MEK Drives BRAF Activation through Allosteric Control of KSR Proteins. Nature 2018, 554, 549–553.
- Yin, X.; Zafrullah, M.; Lee, H.; Haimovitz-Friedman, A.; Fuks, Z.; Kolesnick, R. A Ceramide-Binding C1 Domain Mediates Kinase Suppressor of Ras Membrane Translocation. Cell. Physiol. Biochem. 2009, 24, 219–230.
- Matheny, S.A.; Chen, C.; Kortum, R.L.; Razidlo, G.L.; Lewis, R.E.; White, M.A. Ras Regulates Assembly of Mitogenic Signalling Complexes through the Effector Protein IMP. Nature 2004, 427, 256–260.
- Chen, C.; Lewis, R.E.; White, M.A. IMP Modulates KSR1-Dependent Multivalent Complex Formation to Specify ERK1/2 Pathway Activation and Response Thresholds. J. Biol. Chem. 2008, 283, 12789–12796.
- Matheny, S.A.; White, M.A. Ras-Sensitive IMP Modulation of the Raf/MEK/ERK Cascade through KSR1. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2006; Volume 407, pp. 237–247. ISBN 9780121828127.
- Rajakulendran, T.; Sahmi, M.; Lefrançois, M.; Sicheri, F.; Therrien, M. A Dimerization-Dependent Mechanism Drives RAF Catalytic Activation. Nature 2009, 461, 542–545.
- Verlande, A.; Krafčíková, M.; Potěšil, D.; Trantírek, L.; Zdráhal, Z.; Elkalaf, M.; Trnka, J.; Souček, K.; Rauch, N.; Rauch, J.; et al. Metabolic Stress Regulates ERK Activity by Controlling KSR-RAF Heterodimerization. EMBO Rep. 2018, 19, 320–336.
- Koveal, D.; Schuh-Nuhfer, N.; Ritt, D.; Page, R.; Morrison, D.K.; Peti, W. A CC-SAM, for Coiled Coil-Sterile α Motif, Domain Targets the Scaffold KSR-1 to Specific Sites in the Plasma Membrane. Sci. Signal. 2012, 5, ra94.
- Weissbach, L.; Settleman, J.; Kalady, M.F.; Snijders, A.J.; Murthy, A.E.; Yan, Y.X.; Bernards, A. Identification of a Human RasGAP-Related Protein Containing Calmodulin-Binding Motifs. J. Biol. Chem. 1994, 269, 20517–20521.
- Brill, S.; Li, S.; Lyman, C.W.; Church, D.M.; Wasmuth, J.J.; Weissbach, L.; Bernards, A.; Snijders, A.J. The Ras GTPase-Activating-Protein-Related Human Protein IQGAP2 Harbors a Potential Actin Binding Domain and Interacts with Calmodulin and Rho Family GTPases. Mol. Cell. Biol. 1996, 16, 4869–4878.
- McCallum, S.J.; Wu, W.J.; Cerione, R.A. Identification of a Putative Effector for Cdc42Hs with High Sequence Similarity to the RasGAP-Related Protein IQGAP1 and a Cdc42Hs Binding Partner with Similarity to IQGAP2. J. Biol. Chem. 1996, 271, 21732–21737.
- Wang, S.; Watanabe, T.; Noritake, J.; Fukuta, M.; Yoshimura, T.; Itoh, N.; Harada, T.; Nakagawa, M.; Matsuura, Y.; Arimura, N.; et al. IQGAP3, a Novel Effector of Rac1 and Cdc42, Regulates Neurite Outgrowth. J. Cell Sci. 2007, 120, 567–577.
- Schmidt, V.A.; Scudder, L.; Devoe, C.E.; Bernards, A.; Cupit, L.D.; Bahou, W.F. IQGAP2 Functions as a GTP-Dependent Effector Protein in Thrombin-Induced Platelet Cytoskeletal Reorganization. Blood 2003, 101, 3021–3028.
- Cupit, L.D.; Schmidt, V.A.; Miller, F.; Bahou, W.F. Distinct PAR/IQGAP Expression Patterns Durinq Murine Development: Implications for Thrombin-Associated Cytoskeletal Reorganization. Mamm. Genome 2004, 15, 618–629.
- Nojima, H.; Adachi, M.; Matsui, T.; Okawa, K.; Tsukita, S.; Tsukita, S. IQGAP3 Regulates Cell Proliferation through the Ras/ERK Signalling Cascade. Nat. Cell Biol. 2008, 10, 971–978.
- Li, Z.; Kim, S.H.; Higgins, J.M.G.; Brenner, M.B.; Sacks, D.B. IQGAP1 and Calmodulin Modulate E-Cadherin Function. J. Biol. Chem. 1999, 274, 37885–37892.
- Zhou, R.; Guo, Z.; Watson, C.; Chen, E.; Kong, R.; Wang, W.; Yao, X. Polarized Distribution of IQGAP Proteins in Gastric Parietal Cells and Their Roles in Regulated Epithelial Cell Secretion. Mol. Biol. Cell 2003, 14, 1097–1108.
- Chew, C.S.; Okamoto, C.T.; Chen, X.; Qin, H.Y. IQGAPs Are Differentially Expressed and Regulated in Polarized Gastric Epithelial Cells. Am. J. Physiol.-Gastrointest. Liver Physiol. 2005, 288, G376–G387.
- Hedman, A.C.; Smith, J.M.; Sacks, D.B. The Biology of IQGAP Proteins: Beyond the Cytoskeleton. EMBO Rep. 2015, 16, 427–446.
- Mateer, S.C.; Morris, L.E.; Cromer, D.A.; Benseñor, L.B.; Bloom, G.S. Actin Filament Binding by a Monomeric IQGAP1 Fragment with a Single Calponin Homology Domain. Cell Motil. Cytoskelet. 2004, 58, 231–241.
- Ho, Y.D.; Joyal, J.L.; Li, Z.; Sacks, D.B. IQGAP1 Integrates Ca2+/Calmodulin and Cdc42 Signaling. J. Biol. Chem. 1999, 274, 464–470.
- Li, Z.; Sacks, D.B. Elucidation of the Interaction of Calmodulin with the IQ Motifs of IQGAP1. J. Biol. Chem. 2003, 278, 4347–4352.
- McNulty, D.E.; Li, Z.; White, C.D.; Sacks, D.B.; Annan, R.S. MAPK Scaffold IQGAP1 Binds the EGF Receptor and Modulates Its Activation. J. Biol. Chem. 2011, 286, 15010–15021.
- Roy, M.; Li, Z.; Sacks, D.B. IQGAP1 Is a Scaffold for Mitogen-Activated Protein Kinase Signaling. Mol. Cell. Biol. 2005, 25, 7940–7952.
- Ren, J.G.; Li, Z.; Sacks, D.B. IQGAP1 Modulates Activation of B-Raf. Proc. Natl. Acad. Sci. USA 2007, 104, 10465–10469.
- Jeong, H.W.; Li, Z.; Brown, M.D.; Sacks, D.B. IQGAP1 Binds Rap1 and Modulates Its Activity. J. Biol. Chem. 2007, 282, 20752–20762.
- Ren, J.G.; Li, Z.; Crimmins, D.L.; Sacks, D.B. Self-Association of IQGAP1: Characterization and Functional Sequelae. J. Biol. Chem. 2005, 280, 34548–34557.
- Choi, S.; Thapa, N.; Hedman, A.C.; Li, Z.; Sacks, D.B.; Anderson, R.A. IQGAP1 Is a Novel Phosphatidylinositol 4,5 Bisphosphate Effector in Regulation of Directional Cell Migration. EMBO J. 2013, 32, 2617–2630.
- Ren, J.G.; Li, Z.; Sacks, D.B. IQGAP1 Integrates Ca2+/Calmodulin and B-Raf Signaling. J. Biol. Chem. 2008, 283, 22972–22982.
- Ussar, S.; Voss, T. MEK1 and MEK2, Different Regulators of the G1/S Transition. J. Biol. Chem. 2004, 279, 43861–43869.
- Noritake, J.; Watanabe, T.; Sato, K.; Wang, S.; Kaibuchi, K. IQGAP1: A Key Regulator of Adhesion and Migration. J. Cell Sci. 2005, 118, 2085–2092.
- Le Clainche, C.; Schlaepfer, D.; Ferrari, A.; Klingauf, M.; Grohmanova, K.; Veligodskiy, A.; Didry, D.; Le, D.; Egile, C.; Carlier, M.F.; et al. IQGAP1 Stimulates Actin Assembly through the N-Wasp-Arp2/3 Pathway. J. Biol. Chem. 2007, 282, 426–435.
- Benseñor, L.B.; Kan, H.M.; Wang, N.; Wallrabe, H.; Davidson, L.A.; Cai, Y.; Schafer, D.A.; Bloom, G.S. IQGAP1 Regulates Cell Motility by Linking Growth Factor Signaling to Actin Assembly. J. Cell Sci. 2007, 120, 658–669.
- Kohno, T.; Urao, N.; Ashino, T.; Sudhahar, V.; Inomata, H.; Yamaoka-Tojo, M.; McKinney, R.D.; Fukai, T.; Ushio-Fukai, M. IQGAP1 Links PDGF Receptor-β Signal to Focal Adhesions Involved in Vascular Smooth Muscle Cell Migration: Role in Neointimal Formation after Vascular Injury. Am. J. Physiol.-Cell Physiol. 2013, 305, C591–C600.
- Li, S.; Wang, Q.; Chakladar, A.; Bronson, R.T.; Bernards, A. Gastric Hyperplasia in Mice Lacking the Putative Cdc42 Effector IQGAP1. Mol. Cell. Biol. 2000, 20, 697–701.
- Schmidt, V.A.; Chiariello, C.S.; Capilla, E.; Miller, F.; Bahou, W.F. Development of Hepatocellular Carcinoma in Iqgap2-Deficient Mice Is IQGAP1 Dependent. Mol. Cell. Biol. 2008, 28, 1489–1502.
- Abramovich, C.; Shen, W.F.; Pineault, N.; Imren, S.; Montpetit, B.; Largman, C.; Keith Humphries, R. Functional Cloning and Characterization of a Novel Nonhomeodomain Protein That Inhibits the Binding of PBX1-HOX Complexes to DNA. J. Biol. Chem. 2000, 275, 26172–26177.
- Abramovich, C.; Chavez, E.A.; Lansdorp, P.M.; Humphries, R.K. Functional Characterization of Multiple Domains Involved in the Subcellular Localization of the Hematopoietic Pbx Interacting Protein (Hpip). Oncogene 2002, 21, 6766–6771.
- Manavathi, B.; Acconcia, F.; Rayala, S.K.; Kumar, R. An Inherent Role of Microtubule Network in the Action of Nuclear Receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 15981–15986.
- Khumukcham, S.S.; Manavathi, B. Two Decades of a Protooncogene HPIP/PBXIP1: Uncovering the Tale from Germ Cell to Cancer. Biochim. Biophys. Acta-Rev. Cancer 2021, 1876, 188576.
- Punekar, S.R.; Velcheti, V.; Neel, B.G.; Wong, K.K. The Current State of the Art and Future Trends in RAS-Targeted Cancer Therapies. Nat. Rev. Clin. Oncol. 2022, 19, 637–655.
- Liu, C.; Ye, D.; Yang, H.; Chen, X.; Su, Z.; Li, X.; Ding, M.; Liu, Y. RAS-targeted Cancer Therapy: Advances in Drugging Specific Mutations. MedComm 2023, 4, e285.
- Song, Y.; Bi, Z.; Liu, Y.; Qin, F.; Wei, Y.; Wei, X. Targeting RAS–RAF–MEK–ERK Signaling Pathway in Human Cancer: Current Status in Clinical Trials. Genes Dis. 2023, 10, 76–88.
- Xiao, Y.; Lee, T.; Latham, M.P.; Warner, L.R.; Tanimoto, A.; Pardi, A.; Ahn, N.G. Phosphorylation Releases Constraints to Domain Motion in ERK2. Proc. Natl. Acad. Sci. USA 2014, 111, 2506–2511.
- Anderson, J.W.; Vaisar, D.; Jones, D.N.; Pegram, L.M.; Chen, H.; Moffat, J.G.; Ahn, N.G. Conformation Selection by ATP-Competitive Inhibitors and Allosteric Communication in ERK2. bioRxiv 2023.
- Zhao, L.; Zhao, J.; Zhong, K.; Tong, A.; Jia, D. Targeted Protein Degradation: Mechanisms, Strategies and Application. Signal Transduct. Target. Ther. 2022, 7, 113.
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric Molecules That Target Proteins to the Skp1-Cullin-F Box Complex for Ubiquitination and Degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559.
- Schneekloth, A.R.; Pucheault, M.; Tae, H.S.; Crews, C.M. Targeted Intracellular Protein Degradation Induced by a Small Molecule: En Route to Chemical Proteomics. Bioorg. Med. Chem. Lett. 2008, 18, 5904–5908.
- Lai, A.C.; Toure, M.; Hellerschmied, D.; Salami, J.; Jaime-Figueroa, S.; Ko, E.; Hines, J.; Crews, C.M. Modular PROTAC Design for the Degradation of Oncogenic BCR-ABL. Angew. Chem. Int. Ed. 2016, 55, 807–810.
- Gadd, M.S.; Testa, A.; Lucas, X.; Chan, K.H.; Chen, W.; Lamont, D.J.; Zengerle, M.; Ciulli, A. Structural Basis of PROTAC Cooperative Recognition for Selective Protein Degradation. Nat. Chem. Biol. 2017, 13, 514–521.
- Nalawansha, D.A.; Crews, C.M. PROTACs: An Emerging Therapeutic Modality in Precision Medicine. Cell Chem. Biol. 2020, 27, 998–1014.
- Fang, Y.; Wang, S.; Han, S.; Zhao, Y.; Yu, C.; Liu, H.; Li, N. Targeted Protein Degrader Development for Cancer: Advances, Challenges, and Opportunities. Trends Pharmacol. Sci. 2023, 44, 303–317.
- Mullard, A. Arvinas’s PROTACs Pass First Safety and PK Analysis. Nat. Rev. Drug Discov. 2019, 18, 895.
- Kelm, J.M.; Pandey, D.S.; Malin, E.; Kansou, H.; Arora, S.; Kumar, R.; Gavande, N.S. PROTAC’ing Oncoproteins: Targeted Protein Degradation for Cancer Therapy. Mol. Cancer 2023, 22, 62.
- Zeng, M.; Xiong, Y.; Safaee, N.; Nowak, R.P.; Donovan, K.A.; Yuan, C.J.; Nabet, B.; Gero, T.W.; Feru, F.; Li, L.; et al. Exploring Targeted Degradation Strategy for Oncogenic KRASG12C. Cell Chem. Biol. 2020, 27, 19–31.e6.
- Bond, M.J.; Chu, L.; Nalawansha, D.A.; Li, K.; Crews, C.M. Targeted Degradation of Oncogenic KRASG12Cby VHL-Recruiting PROTACs. ACS Cent. Sci. 2020, 6, 1367–1375.
- Chen, H.; Chen, F.; Pei, S.; Gou, S. Pomalidomide Hybrids Act as Proteolysis Targeting Chimeras: Synthesis, Anticancer Activity and B-Raf Degradation. Bioorg. Chem. 2019, 87, 191–199.
- Posternak, G.; Tang, X.; Maisonneuve, P.; Jin, T.; Lavoie, H.; Daou, S.; Orlicky, S.; Goullet de Rugy, T.; Caldwell, L.; Chan, K.; et al. Functional Characterization of a PROTAC Directed against BRAF Mutant V600E. Nat. Chem. Biol. 2020, 16, 1170–1178.
- Wei, J.; Hu, J.; Wang, L.; Xie, L.; Jin, M.S.; Chen, X.; Liu, J.; Jin, J. Discovery of a First-in-Class Mitogen-Activated Protein Kinase Kinase 1/2 Degrader. J. Med. Chem. 2019, 62, 10897–10911.
- Vollmer, S.; Cunoosamy, D.; Lv, H.; Feng, H.; Li, X.; Nan, Z.; Yang, W.; Perry, M.W.D. Design, Synthesis, and Biological Evaluation of MEK PROTACs. J. Med. Chem. 2020, 63, 157–162.
- Simonetta, K.R.; Taygerly, J.; Boyle, K.; Basham, S.E.; Padovani, C.; Lou, Y.; Cummins, T.J.; Yung, S.L.; von Soly, S.K.; Kayser, F.; et al. Prospective Discovery of Small Molecule Enhancers of an E3 Ligase-Substrate Interaction. Nat. Commun. 2019, 10, 1402.
- Mayor-Ruiz, C.; Bauer, S.; Brand, M.; Kozicka, Z.; Siklos, M.; Imrichova, H.; Kaltheuner, I.H.; Hahn, E.; Seiler, K.; Koren, A.; et al. Rational Discovery of Molecular Glue Degraders via Scalable Chemical Profiling. Nat. Chem. Biol. 2020, 16, 1199–1207.
- Słabicki, M.; Kozicka, Z.; Petzold, G.; Li, Y.D.; Manojkumar, M.; Bunker, R.D.; Donovan, K.A.; Sievers, Q.L.; Koeppel, J.; Suchyta, D.; et al. The CDK Inhibitor CR8 Acts as a Molecular Glue Degrader That Depletes Cyclin K. Nature 2020, 585, 293–297.
- Sasso, J.M.; Tenchov, R.; Wang, D.S.; Johnson, L.S.; Wang, X.; Zhou, Q.A. Molecular Glues: The Adhesive Connecting Targeted Protein Degradation to the Clinic. Biochemistry 2023, 62, 601–623.
- Hao, C.; Li, X.; Wang, Z.; Liu, L.; He, F.; Pan, Z. Optically Activated MEK1/2 Inhibitors (Opti-MEKi) as Potential Antimelanoma Agents. Eur. J. Med. Chem. 2023, 251, 115236.
- Pfaff, P.; Samarasinghe, K.T.G.; Crews, C.M.; Carreira, E.M. Reversible Spatiotemporal Control of Induced Protein Degradation by Bistable PhotoPROTACs. ACS Cent. Sci. 2019, 5, 1682–1690.
