Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Lindsay Dong and Version 1 by Inmaculada Moreno-Cordoba.
There are three concepts linked to the growing resistance to antibiotics, namely (i) the Resistome, which refers to the collection of bacterial genes that confer resistance to antibiotics, (ii) the Mobilome, which includes all the mobile genetic elements that participate in the spreading of antibiotic resistance among bacteria by horizontal gene transfer processes, and (iii) the Nichome, which refers to the set of genes that are expressed when bacteria try to colonize new niches.
- antibiotic resistance
- mobilome
- nichome
- resistome
1. The Antibiotic Resistome
The collection of bacterial genes that can confer single or multiple antimicrobial resistance (AMR) is termed the Resistome [3,16][1][2]. The resistome concept includes: (i) genes involved in resistance to a family of antibiotics (β-lactams, for instance); (ii) genes involved in resistance to multiple antibiotics; (iii) Antibiotic resistance genes (ARGs) belonging to the microbiome of an individual; and (iv) ARGs of bacteria growing in specific niches, like hospitals, soils, or polluted waters [17][3]. A comprehensive antibiotic resistance database, termed CARD, has been updated and curated, and at present permits an in-depth search of the bacterial resistome, although the data are derived from cultivable and pathogenic bacteria [18][4]. An updated version of the previous ResFinder, termed ResFinderFG v2.0 (https://cge.food.dtu.dk/services/ResFinderFG/; accessed on 4 August 2023), which compiles 3913 ARGs identified by functional metagenomics from 50 curated datasets, has been recently released [19][5].
Novel technologies like deep-learning or atom-editing approaches are being used to find new, potentially useful antibiotics [21,22][6][7]. Interestingly, Artificial Intelligence (AI)-based approaches have discovered potentially interesting peptides derived from molecular de-extinction through paleo-proteome mining as a framework for antibacterial drug discovery [23][8]. A database to mine past and present human proteomes to search for potential bioactive encrypted peptides has recently been made available (https://gitlab.com/machine-biology-group-public/pancleave; accessed on 9 August 2023).
The comprehension of infectious diseases caused by bacteria has increased very much but the rapid identification of new bacterial clones, novel mutations, and new variants of pathogens remains a challenge. Further, there are some obscure subjects that we do not fully understand, such as the factors that govern the individual immune response to pathogenic bacteria or how some of these pathogens can evade the immune response of the infected individuals. AMR has been known in bacteria for decades and the present status of a possible solution is still far from satisfactory. Nearly a quarter of a century has elapsed since the beginning of the 21st century, and we continue to be confronted with one of the most important public health issues, which is the increase in AMR over the years [24][9]. The proposed solutions to AMR involve the avoidance of the prescription of antibiotics to treat trivial infections, saving them for more serious ones. A reduction in the total consumption of antibiotics from 2011 to 2022 has been observed, and, despite this, AMR in bacteria has increased over these years, as reported by the European Centre for Disease Prevention and Control in March 2022 (https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-eueea-one-health-response; accessed on 31 July 2023). The accumulated knowledge on the mechanism of action of numerous antibiotics (Table 1, Figure 31) and the realization that several of them cannot be used because of widespread resistance make urgent the prevention of infections and the development of strategies to control them [1][10].
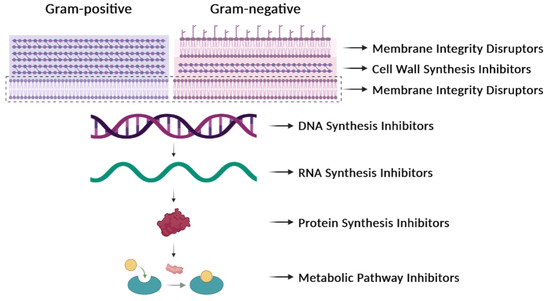
Figure 31. Antibiotic targets. Classification of antibiotics based on their mechanisms of action. They can affect the synthesis of the cell wall, the integrity of the membrane, the synthesis of nucleic acids (DNA or RNA), the synthesis of proteins, or the synthesis of particular metabolites.
Table 1.
Antibiotics: targets and resistance
a
.
| Antibiotic Type: Examples | Targets | Resistance |
|---|---|---|
| β-Lactams: penicillins, cephalosporins | Enzymes involved in cell-wall synthesis | Breakage of the β-lactam ring by β-lactamases. Decreased uptake or increased efflux of the β-lactam. Changes in the active site of the target. |
| Cationic peptides: colistin, polymyxin B | Bacterial membrane | Lipopolysaccharide modifications. Efflux pumps. Capsular polysaccharide production. |
| Fluoroquinolones: ciprofloxacin | DNA gyrase and topoisomerase IV | Mutations in target enzymes. |
| Rifamycins: rifampin, rifabutin, rifapentine | RNA polymerase | Mutations in the RNA polymerase (β subunit). Drug inactivation. Impeded uptake of the drug. |
| Aminoglycosides: streptomycin, gentamicin | 30S ribosomal subunit | Enzymatic modification of the drug. Enzymatic modification of the target site. Efflux pumps. |
| Tetracyclines: tetracycline, doxycycline, minocycline | 30S ribosomal subunit | Efflux pumps. Ribosomal protection proteins. Enzymatic inactivation of tetracycline. |
| Phenicols: chloramphenicol, thiamphenicol, florfenicol | 50S ribosomal subunit | Ribosomal mutations. Enzymatic modification (methylation) of the target site. Enzymatic inactivation (acetylation) of the drug. Efflux pumps. |
| Macrolides: erythromycin, azithromycin, clarithromycin | 50S ribosomal subunit | Ribosomal modification by methylation or mutation. Efflux pumps. Drug inactivation. |
| Lincosamides: clindamycin | 50S ribosomal subunit | Ribosomal modification by methylation or mutation. Efflux pumps. Drug inactivation. |
| Sulfonamides: sulfamethazine, sulfadiazine |
Dihydropteroate synthase in the folic acid pathway | Mutations in the target enzyme. Sulfonamide resistance mediated by HGT (genes encoding variants of the target enzyme). Drug degradation. |
a Further information can be retrieved from the Comprehensive Antibiotic Resistance Database (https://card.mcmaster.ca/; accessed on 31 July 2023).
In general, antibiotics may be grouped by their mechanisms of action (Table 1, Figure 31), which affect: (i) cell-wall synthesis (β-lactams); (ii) protein synthesis (tetracyclines, phenicols, aminoglycosides); (iii) nucleic acid synthesis (rifamycins, fluoroquinolones); and (iv) membrane integrity (polymyxins) [25][11]. Resistance may be due to mutations in the bacterial chromosome or to ARGs carried by MGEs. Frequently, the resistance is the consequence of an enzymatic modification of the antibiotic molecules, like resistance to β-lactams or phenicols. There are several instances in which the resistances are due to Efflux Pumps (EPs), like those involved in resistance to tetracyclines. In several of these cases, a change in the cell morphology concomitant with the onset of antibiotic resistance has been observed [26][12]. EPs are transmembrane transporters able to pump the toxic molecules outside the bacterial cell and can be responsible for resistance to multiple antibiotics (reviewed by [27][13]). Constitutive expression of EP genes is a costly process, causing a heavy genetic burden to the bacteria carrying these genes. However, improved fitness can be achieved by organizing the EP genes in such a way that their mRNAs fold into secondary structures that make the translation inducible by the antibiotic. This is the case for several tet genes (tetracycline-resistance EP genes) [25,28][11][14]. Transcription of the tet genes results in the synthesis of an mRNA that folds into secondary structures that are more or less complex (Figure 42), as it has also been described for some cat (chloramphenicol-acetyl transferases) and erm (erythromycin-methylating enzymes) genes [29][15]. In all these genes, a single promoter directs the synthesis of an mRNA molecule that has two ribosome-binding sites (RBS1 and RBS2) (Figure 42) [30][16].
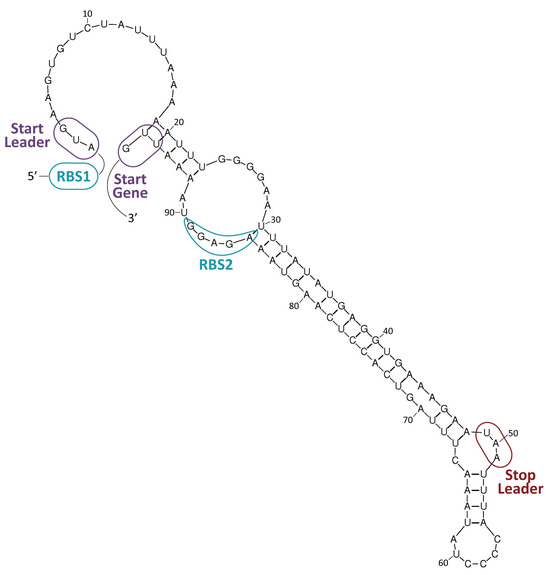

Figure 42. Resistance to tetracycline. The tetK gene of plasmid pT181 (GenBank accession number CP001783.1) encodes a tetracycline-resistance EP [28][14]. A potential secondary structure of its mRNA is shown. Formation of this structure would result in inhibition of translation. RBS1: proposed ribosome-binding site for the translation of a leader peptide from the AUG codon (start leader). RBS2: proposed ribosome-binding site for the translation of the tetK gene from the UUG codon (start gene). The UAA stop codon of the leader peptide is indicated. Expression of the tetK gene is inducible by tetracycline.
2. The Mobilome
The intra- and interspecific spread of antibiotic resistance among bacteria is increased by the existence of ARGs carried by MGEs. The mobile elements may represent up to 25% of the total prokaryotic DNA that is shared among bacteria. The ARGs can be mobilized intracellularly (from chromosome to plasmids and vice versa) or between different bacterial cells [36,37][17][18]. MGEs can be transferred among distantly related bacteria by Horizontal Gene Transfer (HGT) processes, constituting a profusion of circulating DNA termed the Mobilome [38][19]. It includes plasmids, Integrative and Conjugative Elements (ICEs), Integrative and Mobilizable Elements (IMEs), Insertion Sequences (ISs), and prophages and their satellites (Table 2). They all are involved in the transfer of AMR [39,40][20][21]. Nevertheless, the thin red line dividing the different MGEs is, at best, fuzzy because some ICEs and phages do replicate like plasmids, some phages and plasmids integrate into the host chromosome, and some satellites can integrate into phages [40,41][21][22].
Table 2.
Relevant bacterial MGEs.
| Element | Main Features |
|---|---|
| Plasmids | DNA molecules that replicate autonomously. |
| Conjugative Plasmids Mobilizable Plasmids |
Encode all elements needed for conjugation. Encode a relaxase and an origin of transfer. Need the conjugative machinery provided by ‘auxiliary’ plasmids. |
| Integrative and Conjugative Elements (ICEs) | Integrated elements that can excise, replicate, and transfer by conjugation. Previously termed ‘Conjugative Transposons’. |
| Integrative and Mobilizable Elements (IMEs) | Integrated elements that can excise and replicate but need conjugative machinery provided by ‘auxiliary’ elements for their transfer. Previously termed ‘Mobilizable Transposons’. |
| Insertion Sequences (ISs) | Integrated elements that encode only transposition machinery. |
| Prophages | Bacteriophage genomes totally or partially inserted into the host chromosome. |
| Phage satellites | Elements that exploit phages for transfer between bacteria and can encode ARGs. |
2.1. Mobilization Mechanisms
The MGEs spread readily among their bacterial hosts, albeit with different efficiencies. Their dissemination can be achieved by different processes: (i) conjugation, a process that requires contact between donor and recipient cells; (ii) bacteriophage-mediated transduction; and (iii) transformation with free DNA, which occurs in naturally transformable bacteria [14][23]. The three processes may be equally efficient depending on the environment. If the bacterial cells are sessile and live in biofilms, where a matrix of free DNA is generated, natural genetic transformation may be the principal process [42,43,44][24][25][26]. When bacterial cells grow in dynamic environments, such as the mammalian gastrointestinal tract, conjugation could be the main HGT mechanism [45][27]. Furthermore, in bacteria containing prophages, lateral transduction can promote the transfer of large chromosomal regions at high frequencies [46][28]. Nevertheless, the situation may not be as linear as that because it has been shown that: (i) the presence of active prophages can seriously hinder the conjugation rates [47][29], and (ii) interactions between conjugative plasmids and ISs have significantly contributed to the horizontal transfer of AMR genes [48][30]. The classical plasmid conjugation process involves a specific protein (generically termed DNA relaxase), which initiates the transfer by binding to and cleaving at the plasmid origin of transfer, the oriT. The otherwise supercoiled plasmid DNA is relaxed and transferred to the recipient cell in a single-stranded DNA configuration [51][31]. There are, however, MGEs that ‘break the norm’ in the transfer process. There are, for instance, the Streptomyces plasmids, which follow a transfer path more akin to chromosomal segregation than to classical conjugation. The transfer of these plasmids is mediated by the TraB protein through a process that mechanistically resembles DNA translocation during the segregation of daughter molecules at cell division [52,53,54][32][33][34]. Bacteriophage-mediated transduction has been known for decades to be a process to mobilize regions of the bacterial chromosomes [57][35] and small rolling-circle replicating plasmids [58,59][36][37]. More recently, the process termed lateral transduction has been reported, in which not only MGEs but also chromosomal genes are transferred at high frequencies [46][28]. These findings, in conjunction with more recent ones reported by the Penades lab [60[38][39],61], have posed the question of whether the bacterial chromosome could be conceived as an entire MGE, although differences can be conceived in terms of evolutionary selection [62][40]. Natural transformation is defined as the incorporation and integration of pieces of free DNA into the bacterial chromosome by recipient cells that have entered into a specific physiological state termed competence. So far, nearly 100 bacterial species have been shown to undergo this process for genetic variability. In some bacterial species, like A. baumanii (a most relevant member of the ESKAPE group), natural transformation seems to be the most relevant mechanism of HGT, posing the question of whether conjugative transfer is the main HGT process in bacteria [63][41]. In the case of polymyxins (a family of antibiotics that target the cell membrane), it has been speculated that treatment with these antibiotics could lead to the release of free DNA as well as potentiate the onset of the competent state [64][42]. The onset of competence is triggered by a specific sigma factor that turns on a set of operons (generically termed com genes) involved in the active incorporation of free DNA from the environment. The process, in several Gram-positive bacteria, is considered a bi-stable process in which two subpopulations of the bacterial strain differentiate: the non-competent fraction becomes the prey of the competent one, which would commit an act of fratricide or cannibalism [65,66,67,68,69][43][44][45][46][47]. The outbreak of competence was regarded as a survival mechanism in which the competent bacterial population would uptake and integrate some beneficial genetic traits into their chromosome.2.2. Transfer of MGEs
A thorough comparative study has quantified the frequencies of transfer of different MGEs and has reached two interesting conclusions: (i) most of the MGEs participating in HGT belong to the IME category rather than to plasmids, and (ii) less than 1% of the transferred genes are ARGs. Genes conferring other adaptive advantages, like those involved in host invasion or colonization of new niches, were frequently detected [74][48]. It might be argued, however, that HGT can be detected more easily if the gene is integrated into the host chromosome (IMEs, ICEs) than if carried by a plasmid because the latter can be lost in the absence of selective pressure. Thus, many events of plasmid-mediated HGT can occur in nature but only those that spread and remain in the population could be detectable [75][49]. It could also be argued that only plasmids carrying ‘addiction modules’ (i.e., Toxin-Antitoxin modules) that counter-select plasmid-free cells would be the most probable to remain in the population [76][50]. Acquisition of an MGE from the environment usually results in a loss of fitness by the recipient cells (measured as a reduction in the growth rate). Thus, in the absence of selective pressure, MGE-free cells can overgrow those that have recently acquired the MGE [77[51][52][53],78,79], at least until domestication processes take place [80][54].2.3. Inhibition of HGT Processes
Strategies designed to inhibit the transmission of AMR traits by hindering conjugation were proposed early on. A search for Conjugation Inhibitors (COINS) through testing several libraries of natural compounds permitted the selection of some candidates as bona fide lead compounds [82][55]. Additional strategies were published later, like the employment of phage M13 g3p protein [83][56], followed by many interesting proposals to develop new COINS using either natural processes (CRISPR-Cas systems, restriction/modification) or genetically developed processes, like intrabodies against relaxases, obstruction of conjugative pili, or the use of unsaturated fatty acids [84,85,86][57][58][59]. Further, effective inhibition of bacterial conjugative transfer in natural environments has been demonstrated, showing the applicability of particular COINS to effectively block conjugation [87][60]. Additional approaches to reduce the spread of ARGs have been discussed in-depth [6][61]. These include interference with the stability of the mobilome, since by the same token by which MGEs can be acquired, they can be lost. Thus, drugs and processes that activate the loss of MGEs would be an attractive path to explore [88][62]. Another approach contemplates the inhibition of specific systems employed by bacteria to secrete macromolecules, like the T4 Secretion System (T4SS) that participates in conjugation [89][63].3. The Nichome
The bacterial genome has been divided into the ‘core’ and the ‘accessory’ genomes. Whereas the core genome may amount to 20–60% of the bacterial genome, depending upon the species, the accessory genome contains genes that are present in one or more, but not all, strains of the species. These accessory genes might be beneficial when bacteria colonize new niches [97][64]. The mobilome is widely distributed and shared among bacterial species within the accessory genes. Thus, the mobilome can be considered an entity that actively participates in the intra- and interspecific gene spread, leading to an asymmetry in the bacterial populations. The accessory genes carried by the mobilome contribute to the genetic diversity of the bacterial world, and these variations confer advantages when bacteria explore the colonization of new niches [6,98][61][65]. In natural environments, most bacteria live in spatially structured communities composed of several species, the so-called polymicrobial biofilms [99][66]. Under these conditions, the sessile cells within the biofilm may release ‘explorers’ that would search for novel niches where they can either strive and prosper or be eliminated by other competitors or, in the case of infections, by the host immune system [100][67]. In the case of success, cells would colonize the novel niche and would express sets of otherwise silent (or nearly so) genes, the Nichome [6][61]. Genes participating in the nichome may represent an ample amount of the accessory genes, and they are involved in the niche-specific adaptation of the colonizing bacteria [101][68]. When colonizing a new niche, bacteria must be able to sense and respond to the signals that the new niche provides. Thus, the identification of genes responding to these signals will provide precious information on how the whole nichome is regulated. Within the niche, intraspecific signals are released (the quorum sensors, QS) that allow communication between kindred cells, which harbour receptors that respond to the signals released by cells of the same bacterial species, allowing them to cooperate [102][69]. Bacterial adaptation to new niches requires coordination in the expression of numerous genes. This partially relies on regulatory proteins that activate and/or repress the transcription of various genes in response to environmental stimuli (global transcriptional regulators). Among them, (i) regulatory proteins that are associated with a transmembrane sensor histidine kinase, the so-called two-component signal transduction systems (TCSs); (ii) nucleoid-associated proteins (NAPs) that play an essential role not only in maintaining the DNA architecture but also in regulating gene expression; and (iii) regulatory proteins of the Mga/AtxA family, which has been defined as a new class of regulators containing phosphoenolpyruvate:carbohydrate phosphotransferase system (PTS) regulation domains. TCSs are widespread in bacteria but absent in animals and humans. Many studies illustrate their important roles in bacterial pathogenesis and antibiotic resistance [108,109,110][70][71][72]. The molecular mechanisms involved in the response to environmental changes by TCSs have been studied in detail [111][73]. In a prototypical TCS, the regulator generates a specific cellular response to the signal detected by the cognate histidine kinase. Most of the classified response regulators, but not all of them, contain a DNA-binding domain and are expected to function as transcriptional regulators. Generally, phosphorylation of this class of regulators by the histidine kinase increases the affinity of the regulator for specific DNA motifs, usually direct or inverted repeats. Thus, the activated regulator binds to its target promoters and alters (activates or represses) gene expression. Proteins that can modulate the activity of particular TCSs by interacting with either the histidine kinase or the response regulator have been identified [112,113,114][74][75][76]. In some cases, such proteins establish connections between TCSs and enable one system to respond to the signal detected by a different system [115][77]. NAPs are architectural proteins of the bacterial chromosome. It has been described that some of them participate in AMR [117][78]. Most of the NAPs play key roles as global regulators of gene expression, as is the case of H-NS (histone-like nucleoid structuring protein) and HU from enterobacteria [118][79]. H-NS functions generally as a transcriptional repressor. It regulates a wide range of genes in response to changes in osmolarity, pH, or temperature [119,120,121][80][81][82]. In vitro, DNA binding experiments have shown that H-NS does not recognize particular DNA sequences, but it has a strong preference for intrinsically curved AT-rich DNA regions [122][83]. Moreover, structural studies have shown that the DNA-binding domain of Ler, a member of the H-NS protein family, does not participate in base-specific contacts but recognizes specific structural features in the DNA minor groove [123][84]. In solution, H-NS can form higher-order oligomers, which correlates with its ability to form nucleoprotein filaments, either linear filaments (binding of H-NS to one DNA duplex) or bridged filaments (binding of H-NS to two segments of DNA duplex). Both types of nucleoprotein filaments can suppress transcription by different mechanisms [120,121,124,125,126,127][81][82][85][86][87][88]. It has been shown that H-NS selectively silences the expression of AT-rich horizontally acquired genes, including pathogenicity islands. This silencing is crucial during the integration of foreign genes into the host genome to ensure that bacterial fitness is maintained [128,129][89][90]. Proteins that can enhance H-NS-mediated silencing have been identified (e.g., Hha and StpA) [130,131][91][92]. As well as H-NS, the bacterial NAP termed HU plays a critical role in global gene regulation. In E. coli, the HU regulon includes genes involved in the adaptive response to anaerobiosis, acid stress, and high osmolarity [137,138][93][94]. HU binds to double-stranded DNA with low affinity and without sequence specificity. However, it binds specifically and with high affinity to defined DNA structures [139,140,141][95][96][97]. Studies on the transcription of the gal operon of E. coli showed that its repression by GalR and HU requires a structure-specific HU binding [142][98]. In the case of Streptococcus pneumoniae, the SpnHU protein, which seems to be the only NAP encoded by this bacterium, was shown to be essential to maintaining the superhelicity of its chromosome [143][99].References
- Hobson, C.; Chan, A.N.; Wright, G.D. The antibiotic resistome: A guide for the discovery of natural products as antimicrobial agents. Chem. Rev. 2021, 121, 3464–3494.
- Wright, G.D. The antibiotic resistome: The nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007, 5, 175–186.
- Kunhikannan, S.; Thomas, C.J.; Franks, A.E.; Mahadevaiah, S.; Kumar, S.; Petrovski, S. Environmental hotspots for antibiotic resistance genes. Microbiologyopen 2021, 10, e1197.
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2022, 51, D690–D699.
- Gschwind, R.; Ugarcina Perovic, S.; Weiss, M.; Petitjean, M.; Lao, J.; Coelho, L.P.; Ruppé, E. ResFinderFG v2.0: A database of antibiotic resistance genes obtained by functional metagenomics. Nucleic Acids Res. 2023, 51, W493–W500.
- Liu, G.; Catacutan, D.B.; Rathod, K.; Swanson, K.; Jin, W.; Mohammed, J.C.; Chiappino-Pepe, A.; Syed, S.A.; Fragis, M.; Rachwalski, K.; et al. Deep learning-guided discovery of an antibiotic targeting Acinetobacter baumannii. Nat. Chem. Biol. 2023.
- Peplow, M. Skeleton crew. Nature 2023, 618, 21–24.
- Maasch, J.R.M.A.; Torres, M.D.T.; Melo, M.C.R.; de la Fuente-Nunez, C. Molecular de-extinction of ancient antimicrobial peptides enabled by machine learning. Cell Host Microbe 2023, 31, 1260–1274.e6.
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Virulence Mech. Bact. Pathog. 2016, 4, 481–511.
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80.
- Sultan, I.; Rahman, S.; Jan, A.T.; Siddiqui, M.T.; Mondal, A.H.; Haq, Q.M.R. Antibiotics, resistome and resistance mechanisms: A bacterial perspective. Front. Microbiol. 2018, 9, 2066.
- Ojkic, N.; Serbanescu, D.; Banerjee, S.; Harwood, C.S. Antibiotic resistance via bacterial cell shape-shifting. mBio 2022, 13, e00659-22.
- Pasqua, M.; Bonaccorsi di Patti, M.C.; Fanelli, G.; Utsumi, R.; Eguchi, Y.; Trirocco, R.; Prosseda, G.; Grossi, M.; Colonna, B. Host-Bacterial pathogen communication: The wily role of the multidrug efflux pumps of the MFS family. Front. Mol. Biosci. 2021, 8, 723274.
- Guay, G.G.; Khan, S.A.; Rothstein, D.M. The tet(K) gene of plasmid pT181 of Staphylococcus aureus encodes an efflux protein that contains 14 transmembrane helices. Plasmid 1993, 30, 163–166.
- Rowe, S.J.; Mecaskey, R.J.; Nasef, M.; Talton, R.C.; Sharkey, R.E.; Halliday, J.C.; Dunkle, J.A. Shared requirements for key residues in the antibiotic resistance enzymes ErmC and ErmE suggest a common mode of RNA recognition. J. Biol. Chem. 2020, 295, 17476–17485.
- Osterman, I.A.; Chervontseva, Z.S.; Evfratov, S.A.; Sorokina, A.V.; Rodin, V.A.; Rubtsova, M.P.; Komarova, E.S.; Zatsepin, T.S.; Kabilov, M.R.; Bogdanov, A.A.; et al. Translation at first sight: The influence of leading codons. Nucleic Acids Res. 2020, 48, 6931–6942.
- Thomas, C.M. The Horizontal Gene Pool; Harwood Academic Publishers: Amsterdam, The Netherlands, 2000; p. 419.
- Dimitriu, T. Evolution of horizontal transmission in antimicrobial resistance plasmids. Microbiology 2022, 168, 1214.
- Siefert, J.L. Defining the mobilome. Methods Mol. Biol. 2009, 532, 13–27.
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17.
- De Sousa, J.A.M.; Fillol-Salom, A.; Penadés, J.R.; Rocha, E.P.C. Identification and characterization of thousands of bacteriophage satellites across bacteria. Nucleic Acids Res. 2023, 51, 2759–2777.
- Carr, V.R.; Shkoporov, A.; Hill, C.; Mullany, P.; Moyes, D.L. Probing the Mobilome: Discoveries in the dynamic microbiome. Trends Microbiol. 2021, 29, 158–170.
- Garcillán-Barcia, M.P.; Pluta, R.; Lorenzo-Díaz, F.; Bravo, A.; Espinosa, M. The facts and family secrets of plasmids that replicate via the rolling-circle mechanism. Microbiol. Mol. Biol. Rev. 2022, 86, e00222-20.
- Domenech, M.; Ruiz, S.; Moscoso, M.; García, E. In vitro biofilm development of Streptococcus pneumoniae and formation of choline-binding protein–DNA complexes. Environ. Microbiol. Rep. 2015, 7, 715–727.
- Carvalho, G.; Fouchet, D.; Danesh, G.; Godeux, A.-S.; Laaberki, M.-H.; Pontier, D.; Charpentier, X.; Venner, S. Bacterial transformation buffers environmental fluctuations through the reversible integration of mobile genetic elements. mBio 2020, 11, e02443-19.
- Michaelis, C.; Grohmann, E. Horizontal Gene Transfer of antibiotic resistance genes in biofilms. Antibiotics 2023, 12, 328.
- Garcia-Quintanilla, M.; Ramos-Morales, F.; Casadesus, J. Conjugal Transfer of the Salmonella enterica Virulence Plasmid in the Mouse Intestine. J. Bacteriol. 2008, 190, 1922–1927.
- Chen, J.; Quiles-Puchalt, N.; Chiang, Y.N.; Bacigalupe, R.; Fillol-Salom, A.; Chee, M.S.J.; Fitzgerald, J.R.; Penadés, J.R. Genome hypermobility by lateral transduction. Science 2018, 362, 207.
- Igler, C.; Schwyter, L.; Gehrig, D.; Wendling, C.C. Conjugative plasmid transfer is limited by prophages but can be overcome by high conjugation rates. Philos. Trans. R. Soc. B Biol. Sci. 2022, 377, 20200470.
- Che, Y.; Yang, Y.; Xu, X.; Břinda, K.; Polz, M.F.; Hanage, W.P.; Zhang, T. Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc. Natl. Acad. Sci. USA 2021, 118, e2008731118.
- Cabezón, E.; Ripoll-Rozada, J.; Peña, A.; de la Cruz, F.; Arechaga, I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2015, 39, 81–95.
- Thoma, L.; Dobrowinski, H.; Finger, C.; Guezguez, J.; Linke, D.; Sepulveda, E.; Muth, G. A multiprotein DNA translocation complex directs intramycelial plasmid spreading during Streptomyces conjugation. mBio 2015, 6, e02559-14.
- Thoma, L.; Muth, G. Conjugative DNA transfer in Streptomyces by TraB: Is one protein enough? FEMS Microbiol. Lett. 2012, 337, 81–88.
- Thoma, L.; Muth, G. Conjugative DNA-transfer in Streptomyces, a mycelial organism. Plasmid 2016, 87–88, 1–9.
- Tyler, B.M.; Goldberg, R.B. Transduction of chromosomal genes between enteric bacteria by bacteriophage P1. J. Bacteriol. 1976, 125, 1105–1111.
- Deichelbohrer, I.; Alonso, J.C.; Lüder, G.; Trautner, T.A. Plasmid transduction by Bacillus subtilis bacteriophage SPP1: Effects of DNA homology between plasmid and bacteriophage. J. Bacteriol. 1985, 162, 1238–1243.
- Novick, R.P.; Edelman, I.; Lofdahl, S. Small Staphylococcus aureus plasmids are transduced as linear multimers that are formed and resolved by replicative processes. J. Mol. Biol. 1986, 192, 209–220.
- Brady, A.; Felipe-Ruiz, A.; Gallego del Sol, F.; Marina, A.; Quiles-Puchalt, N.; Penadés, J.R. Molecular Basis of Lysis–Lysogeny Decisions in Gram-Positive Phages. Annu. Rev. Microbiol. 2021, 75, 563–581.
- Humphrey, S.; Fillol-Salom, A.; Quiles-Puchalt, N.; Ibarra-Chávez, R.; Haag, A.F.; Chen, J.; Penadés, J.R. Bacterial chromosomal mobility via lateral transduction exceeds that of classical mobile genetic elements. Nat. Commun. 2021, 12, 6509.
- Hall, J.P.J. Is the bacterial chromosome a mobile genetic element? Nat. Commun. 2021, 12, 6400.
- Godeux, A.-S.; Svedholm, E.; Barreto, S.; Potron, A.; Venner, S.; Charpentier, X.; Laaberki, M.-H.; Bonomo Robert, A. Interbacterial transfer of carbapenem resistance and large antibiotic resistance islands by natural transformation in pathogenic Acinetobacter. mBio 2022, 13, e02631-21.
- Perez, F.; Stiefel, U. The impact of natural transformation on the acquisition of antibiotic resistance determinants. mBio 2022, 13, e00336-22.
- Claverys, J.P.; Havarstein, L.S. Cannibalism and fratricide: Mechanisms and raisons d’être. Nat. Rev. Microbiol. 2007, 5, 219–229.
- Dubnau, D.; Blokesch, M. Mechanisms of DNA uptake by naturally competent bacteria. Annu. Rev. Genet. 2019, 53, 217–237.
- Havarstein, L.S.; Martin, B.; Johnsborg, O.; Granadel, C.; Claverys, J.P. New insights into pneumococcal fratricide: Relationship to clumping and identification of a novel immunity factor. Mol. Microbiol. 2006, 59, 1297–1307.
- Dubnau, D.; Losick, R. Bistability in bacteria. Mol. Microbiol. 2006, 61, 564–572.
- González-Pastor, J.E.; Hobbs, E.C.; Losick, R. Cannibalism by sporulating bacteria. Science 2003, 301, 510–513.
- Khedkar, S.; Smyshlyaev, G.; Letunic, I.; Maistrenko, O.M.; Coelho, L.P.; Orakov, A.; Forslund, S.K.; Hildebrand, F.; Luetge, M.; Schmidt, T.S.B.; et al. Landscape of mobile genetic elements and their antibiotic resistance cargo in prokaryotic genomes. Nucleic Acids Res. 2022, 50, 3155–3168.
- Martínez, J.L.; Coque, T.M.; Lanza, V.F.; de la Cruz, F.; Baquero, F. Genomic and metagenomic technologies to explore the antibiotic resistance mobilome. Ann. N. Y. Acad. Sci. 2017, 1388, 26–41.
- Díaz-Orejas, R.; Espinosa, M.; Yeo, C.C. The importance of the expendable: Toxin–Antitoxin genes in plasmids and chromosomes. Front. Microbiol. 2017, 8, 1479.
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010, 8, 260–271.
- Martínez, J.L.; Baquero, F.; Andersson, D.I. Beyond serial passages: New methods for predicting the emergence of resistance to novel antibiotics. Curr. Opin. Pharmacol. 2011, 11, 439–445.
- Hernández-Arriaga, A.M.; Espinosa, M.; del Solar, G. Fitness of the pMV158 replicon in Streptococcus Pneumoniae. Plasmid 2012, 67, 162–166.
- Touchon, M.; Bobay, L.-M.; Rocha, E.P.C. The chromosomal accommodation and domestication of mobile genetic elements. Curr. Opin. Microbiol. 2014, 22, 22–29.
- Fernandez-Lopez, R.; Machon, C.; Longshaw, C.M.; Martin, S.; Molin, S.; Zechner, E.L.; Espinosa, M.; Lanka, E.; de la Cruz, F. Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology 2005, 151, 3517–3526.
- Lin, A.; Jimenez, J.; Derr, J.; Vera, P.; Manapat, M.L.; Esvelt, K.M.; Villanueva, L.; Liu, D.R.; Chen, I.A. Inhibition of bacterial conjugation by phage M13 and its protein g3p: Quantitative analysis and model. PLoS ONE 2011, 6, e19991.
- Cabezón, E.; de la Cruz, F.; Arechaga, I. Conjugation inhibitors and their potential use to prevent dissemination of antibiotic resistance genes in bacteria. Front. Microbiol. 2017, 8, 2329.
- Garcillán-Barcia, M.P.; Jurado, P.; González-Pérez, B.; Moncalián, G.; Fernández, L.A.; de la Cruz, F. Conjugative transfer can be inhibited by blocking relaxase activity within recipient cells with intrabodies. Mol. Microbiol. 2007, 63, 404–416.
- Getino, M.; de la Cruz, F. Natural and artificial strategies to control the conjugative transmission of plasmids. Microbiol. Spectr. 2018, 6, 10-1128.
- Palencia-Gándara, C.; Getino, M.; Moyano, G.; Redondo, S.; Fernández-López, R.; González-Zorn, B.; de la Cruz, F. Conjugation inhibitors effectively prevent plasmid transmission in natural environments. mBio 2021, 12, e01277-21.
- Bravo, A.; Ruiz-Cruz, S.; Alkorta, I.; Espinosa, M. When humans met superbugs: Strategies to tackle bacterial resistances to antibiotics. BMC 2018, 9, 216–226.
- Vrancianu, C.O.; Popa, L.I.; Bleotu, C.; Chifiriuc, M.C. Targeting plasmids to limit acquisition and transmission of antimicrobial resistance. Front. Microbiol. 2020, 11, 761.
- Álvarez-Rodríguez, I.; Arana, L.; Ugarte-Uribe, B.; Gómez-Rubio, E.; Martín-Santamaría, S.; Garbisu, C.; Alkorta, I. Type IV Coupling Proteins as potential targets to control the dissemination of antibiotic resistance. Front. Mol. Biosci. 2020, 7, 201.
- Rouli, L.; Merhej, V.; Fournier, P.E.; Raoult, D. The bacterial pangenome as a new tool for analysing pathogenic bacteria. New Microbes New Infect. 2015, 7, 72–85.
- Vernikos, G.; Medini, D.; Riley, D.R.; Tettelin, H. Ten years of pan-genome analyses. Curr. Opin. Microbiol. 2015, 23, 148–154.
- Flemming, H.-C.; Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260.
- Sempere, J.; Llamosí, M.; Román, F.; Lago, D.; González-Camacho, F.; Pérez-García, C.; Yuste, J.; Domenech, M. Clearance of mixed biofilms of Streptococcus pneumoniae and methicillin-susceptible/resistant Staphylococcus aureus by antioxidants N-acetyl-l-cysteine and cysteamine. Sci. Rep. 2022, 12, 6668.
- Douglas, G.M.; Shapiro, B.J. Genic selection within prokaryotic pangenomes. Genome Biol. Evol. 2021, 13, evab234.
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320.
- Tierney, A.R.P.; Rather, P.N. Roles of two-component regulatory systems in antibiotic resistance. Future Microbiol. 2019, 14, 533–552.
- Bhagirath, A.Y.; Li, Y.; Patidar, R.; Yerex, K.; Ma, X.; Kumar, A.; Duan, K. Two component regulatory systems and antibiotic resistance in Gram-negative pathogens. Int. J. Mol. Sci. 2019, 20, 1781.
- Stupar, M.; Furness, J.; De Voss, C.J.; Tan, L.; West, N.P. Two-component sensor histidine kinases of Mycobacterium tuberculosis: Beacons for niche navigation. Mol. Microbiol. 2022, 117, 973–985.
- Zschiedrich, C.P.; Keidel, V.; Szurmant, H. Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 2016, 428, 3752–3775.
- Kato, A.; Groisman, E.A. Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes Dev. 2004, 18, 2302–2313.
- Firon, A.; Tazi, A.; Da Cunha, V.; Brinster, S.; Sauvage, E.; Dramsi, S.; Golenbock, D.T.; Glaser, P.; Poyart, C.; Trieu-Cuot, P. The Abi-domain protein Abx1 interacts with the CovS histidine kinase to control virulence gene expression in group B Streptococcus. PLoS Pathog. 2013, 9, e1003179.
- Mitchell, S.L.; Ismail, A.M.; Kenrick, S.A.; Camilli, A. The VieB auxiliary protein negatively regulates the VieSA signal transduction system in Vibrio cholerae. BMC Microbiol. 2015, 15, 59.
- Mitrophanov, A.Y.; Groisman, E.A. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008, 22, 2601–2611.
- Wang, X.; Yu, D.; Chen, L. Antimicrobial resistance and mechanisms of epigenetic regulation. Front. Cell. Infect. Microbiol. 2023, 13, 1199646.
- Dillon, S.C.; Dorman, C.J. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010, 8, 185–195.
- Atlung, T.; Ingmer, H. H-NS: A modulator of environmentally regulated gene expression. Mol. Microbiol. 1997, 24, 7–17.
- Amit, R.; Oppenheim, A.B.; Stavans, J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys. J. 2003, 84, 2467–2473.
- Liu, Y.; Chen, H.; Kenney, L.J.; Yan, J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010, 24, 339–344.
- Winardhi, R.S.; Yan, J.; Kenney, L.J. H-NS regulates gene expression and compacts the nucleoid: Insights from single-molecule experiments. Biophys. J. 2015, 109, 1321–1329.
- Cordeiro, T.N.; Schmidt, H.; Madrid, C.; Juárez, A.; Bernadó, P.; Griesinger, C.; García, J.; Pons, M. Indirect DNA readout by an H-NS related protein: Structure of the DNA complex of the C-terminal domain of Ler. PLoS Pathog. 2011, 7, e1002380.
- Dame, R.T.; Wyman, C.; Wurm, R.; Wagner, R.; Goosen, N. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1 J. Biol. Chem. 2002, 277, 2146–2150.
- Lim, C.J.; Lee, S.Y.; Kenney, L.J.; Yan, J. Nucleoprotein filament formation is the structural basis for bacterial protein H-NS gene silencing. Sci. Rep. 2012, 2, 509.
- Singh, S.S.; Singh, N.; Bonocora, R.P.; Fitzgerald, D.M.; Wade, J.T.; Grainger, D.C. Widespread suppression of intragenic transcription initiation by H-NS. Genes Dev. 2014, 28, 214–219.
- Kotlajich, M.V.; Hron, D.R.; Boudreau, B.A.; Sun, Z.; Lyubchenko, Y.L.; Landick, R. Bridged filaments of histone-like nucleoid structuring protein pause RNA polymerase and aid termination in bacteria. eLife 2015, 4, e04970.
- Navarre, W.W.; Porwollik, S.; Wang, Y.; McClelland, M.; Rosen, H.; Libby, S.J.; Fang, F.C. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 2006, 313, 236–238.
- Lucchini, S.; Rowley, G.; Goldberg, M.D.; Hurd, D.; Harrison, M.; Hinton, J.C.D. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006, 2, e81.
- Ali, S.S.; Whitney, J.C.; Stevenson, J.; Robinson, H.; Howell, P.L.; Navarre, W.W. Structural insights into the regulation of foreign genes in Salmonella by the Hha/H-NS complex. J. Biol. Chem. 2013, 288, 13356–13369.
- Boudreau, B.A.; Hron, D.R.; Qin, L.; van der Valk, R.A.; Kotlajich, M.V.; Dame, R.T.; Landick, R. StpA and Hha stimulate pausing by RNA polymerase by promoting DNA-DNA bridging of H-NS filaments. Nucleic Acids Res. 2018, 46, 5525–5546.
- Oberto, J.; Nabti, S.; Jooste, V.; Mignot, H.; Rouviere-Yaniv, J. The HU regulon is composed of genes responding to anaerobiosis, acid stress, high osmolarity and SOS induction. PLoS ONE 2009, 4, e4367.
- Prieto, A.I.; Kahramanoglou, C.; Ali, R.M.; Fraser, G.M.; Seshasayee, A.S.N.; Luscombe, N.M. Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 2012, 40, 3524–3537.
- Pinson, V.; Takahashi, M.; Rouviere-Yaniv, J. Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA J. Mol. Biol. 1999, 287, 485–497.
- Kamashev, D.; Balandina, A.; Rouviere-Yaniv, J. The binding motif recognized by HU on both nicked and cruciform DNA. EMBO J. 1999, 18, 5434–5444.
- Kamashev, D.; Rouviere-Yaniv, J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. EMBO J. 2000, 19, 6527–6535.
- Lyubchenko, Y.L.; Shlyakhtenko, L.S.; Aki, T.; Adhya, S. Atomic force microscopic demonstration of DNA looping by GalR and HU. Nucleic Acids Res. 1997, 25, 873–876.
- Ferrándiz, M.-J.; Carreño, D.; Ayora, S.; de la Campa, A.G. HU of Streptococcus pneumoniae is essential for the preservation of DNA supercoiling. Front. Microbiol. 2018, 9, 493.
More
