Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Jessie Wu and Version 3 by Jessie Wu.
Hepatitis B virus (HBV) and its close relative woodchuck hepatitis virus (WHV) belong to the hepadnaviral family. Both are highly oncogenic DNA viruses, and their persistent infection and integration into the host’s hepatocyte genome are the main contributors to the development of hepatocellular carcinoma (HCC).
- HBV
- woodchuck hepatitis virus
- early hepadnavirus–host DNA integration
- virus-induced oxidative DNA damage
1. Hepadnavirus DNA Breaking Points Yielding Early Fusions with Hepatocyte Genome
The localization of hepatitis B virus (HBV) and hepatitis virus (WHV) DNA breakpoints creating junctions with host sequences in the first hours after exposure to the virus was limited to the X gene sequence owing to the design of the primers applied for inv-PCR amplifications. Undeniably, DNA breaks also occur in other parts of HBV and WHV genomes, and they can create merges in the initial stages of infection. This was already exemplified when primers for the WHV preS genomic region were used to identify WHV DNA insertions and breakpoints in the preS and P gene sequences in liver biopsies acquired from woodchucks at 1 and 3 h p.i. (see Figure 1) [1].

Figure 1. Examples of the earliest HBV and WHV DNA fusions with human or woodchuck genomic sequences detected between 15 min and 1 h after exposure to virus in different infection systems investigated in researchers' studies. With the exception of the overlapping homologous junction (OHJ) created by the HBV X gene (HBV X) and human neurotrimin (NTM) gene displayed on the left side of the top panel, all other virus–host genomic fusions shown have the format of the head-to-tail junction (HTJ). In the bottom left panel, the virus–host DNA merge was formed by the preS sequence of WHV S (envelope) gene (WHV PS) and it was detected in a liver biopsy obtained from a woodchuck one hour after injection with WHV, as described in the text. Other abbreviations: SINE, short-interspersed nuclear element (retrotransposon); MAML-2, mastermind-like 2; WHV X, WHV X gene; host, unidentified woodchuck genomic sequence.
Taking into consideration the data from HepaRG and HepG2-NTCP-C4 cells obtained up to one hour after exposure to the virus, there were eight HBV breakpoints found in total [1][2]. Six of them occurred within the X gene fragment overlapping the Enh-II sequence, and the remaining two occurred in the base core region (BCR) of the core promoter (Figure 2). There were also 10 other breakpoints detected at 3 h and 24 h after infection. Only one of them, detected at 3 h p.i., was located within Enh-II; three were detected in the upstream regulatory region (URR) of the core promoter just before the Enh-II sequence, three more were detected within the BCR, two were detected at the beginning of the X gene sequence, and one was detected outside the X gene at the beginning of the core gene (Figure 2). By analyzing the data reported by another group, which were collected at 24 h after the infection of Huh7-NTCP cells with HBV [3], researchers found that one of the four HBV breakpoints was within the Enh-II sequence, and the remaining three were within the BCR. Overall, the results gave the impression that the earliest integration sites identified at 30 min and 1 h p.i. were created mainly by the breakpoints emerging in the HBV Enh-II sequence (Figure 2). By contrast, the BCP appeared to be the most prone to DNA breakages one hour after exposure to the virus, i.e., at 3 h and 24 h p.i. This might be an interesting observation since both Enh-II and the BCR are important regulatory elements relevant to HBV replication, and they may also modulate the expression of the merged host sequences. The analysis of more breakpoints within these elements and the recognition of the transcriptional activity and functional consequences of the resulting chimeras are required before this possibility becomes less elusive. WHV DNA breaks forming junctions with woodchuck genomes in liver biopsies collected in the first 3 h p.i. were also predominantly located in the WHx gene fragment overlapping the WHV BCP sequence, and five different breakpoints were found at this location [1]. In WCM260 hepatocytes exposed to WHV, virus integrations were discovered as early as 15 min p.i., and all WHV breakpoints found at this time point were located within the virus BCP sequence (Chauhan and Michalak, data unpublished).
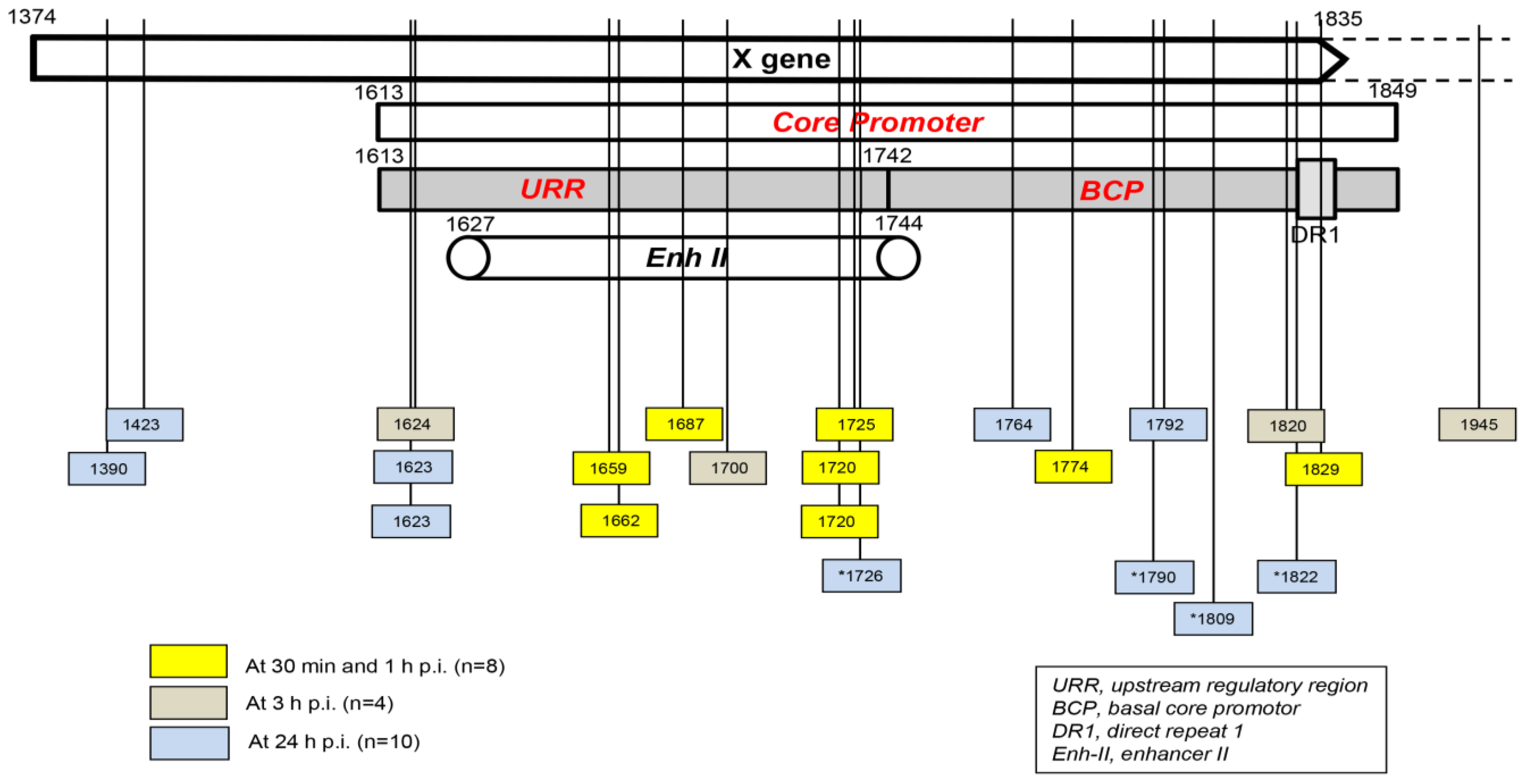
Figure 2. Schematic presentation of the HBV X gene break points forming fusions with the human genomic sequences that were detected in hepatocyte-compatible HepRG and HepG2-NRCP-C4 cells within the first 24 h after exposure to HBV. Yellow squares show breakpoints that formed junctions detected at 30 min and 1 h after exposure to virus, red squares show those identified at 3 h post-infection, and blue squares show those detected at 24 h post-infection. Blue squares with stars show breakpoints reported at 24 h after infection in reference [3]. Numbers mark nucleotide positions according to the HBV DNA GenBank X79185 sequence. Abbreviations: URR, upstream regulatory region of the HBV core promoter; BCP, basal core promoter; DR1, direct repeat region; Enh II, HBV enhancer II.
2. Molecular Format of the Earliest Hepadnavirus–Host Genomic Junctions
The great majority of the HBV– and WHV–host genomic fusions detected in the first hour p.i. were merges of the HTJ type. By contrast, those with the OHJ format were very rare (Figure 2) [1][2][4]. Thus, among 8 HBV and 19 WHV insertional sites identified during this time period, only one (3.7%) was of the OHJ type, and it was created by HBV and the NTM gene sequence, as mentioned above [1]. The same situation was seen when the host sites of HBV or WHV insertions were enumerated between one and 24 h p.i. Thus, a further nine HBV and three WHV sites were identified. Among them, again, only one merge displayed the OHJ format, implying that it was created by micro-homology overlapping joining (MHOJ). This particular fusion was formed by HBV and the runt-related transcription factor 1 (RunX1) sequence at 24 h p.i. in HepG2-NTCP-C4 cells [2]. The HTJ formats of four other HBV–host fusions were identified in Huh7-NTCP cells exposed to recombinant HBV and investigated at 24 h p.i. by another group [3]. Taken together, the results showed that HBV and WHV, in the earliest stages of infection, are joined with hepatocyte genomic sequences almost entirely via HTJs. The creation of such junctions reflects the involvement of the non-homologous end-joining (NHEJ) mechanism [5][6]. Directed by this finding and by the fact that NHEJ is involved in the repair of cell dsDNA breaks, the presence of which is a prerequisite for HBV DNA integration [7], researchers wanted to identify a molecular thread connecting these events.
3. Mechanism of Formation of the Initial Hepadnavirus–Host Genomic Merges
It was shown that HBV could induce oxidative stress by triggering the production of reactive oxygen species (ROS) and reactive nitrogen species (RNS) that cause cell DNA oxidation and oxidation-induced double-stranded (ds) DNA breakages [8]. To assess whether oxidative DNA damage contributes to the formation of the initial hepadnavirus–host DNA junctions, woodchuck WCM260 hepatocytes in which WHV–host HTJs were formed from 15 min p.i. were investigated from this time point onward. For details on the kinetics of the individual markers of oxidative stress, cell DNA damage, and DNA repair, please see Figures 2–5 included in reference [4]. Thus, researchers found a strong and prolonged induction of ROS but only very transient production of inducible nitric oxide (iNOS), which both became detectable from 15 min after exposure to the virus (Figure 3). Notably, while ROS reactivity remained highly elevated between 15 min and 6 h after infection, the activity of iNOS increased only briefly between 15 and 30 min p.i. (see Figure 2 in reference [4]). This coincided with microscopically evident cellular DNA damage, which significantly increased between 15 min and 1 h p.i., as determined by single-cell alkaline comet assay and the nuclear tail length measurements (see Figure 2 in reference [4]). To determine whether the repair of the DNA breakages due to virus-induced oxidative stress was responsible for the formation of the initial virus–host fusions, the time kinetics of the transcription of poly(ADP-ribose) polymerase 1 (PARP1), which recognizes dsDNA breaks and facilitates their repair by the alternative NHEJ pathway, and X-ray repair cross-complementing protein 1 (XRCC1), which is a binding partner of PARP1 in this process, were quantified (see Figure 3Figure 4 in reference [4]). This was supplemented by the quantification of nicotinamide adenine dinucleotide (NAD+) activity, an indicator of PARP1 activation; the transcription of heme oxygenase-1 (HO1), a marker of pro-oxidative cell stress; the transcription of 8-oxyguanidine DNA glucose 1 (OGG1), an indicator of cell response to oxidative DNA damage; and the quantification of the PARP1 cleavage activity (see Figures 4 and 5 shown in reference [4]). The data revealed the synchronized induction of PARP1 and XRCC1 transcription accompanied by augmented reactivity of NAD+ and HO1, which all were initiated at 15 to 30 min p.i. (Figure 3).
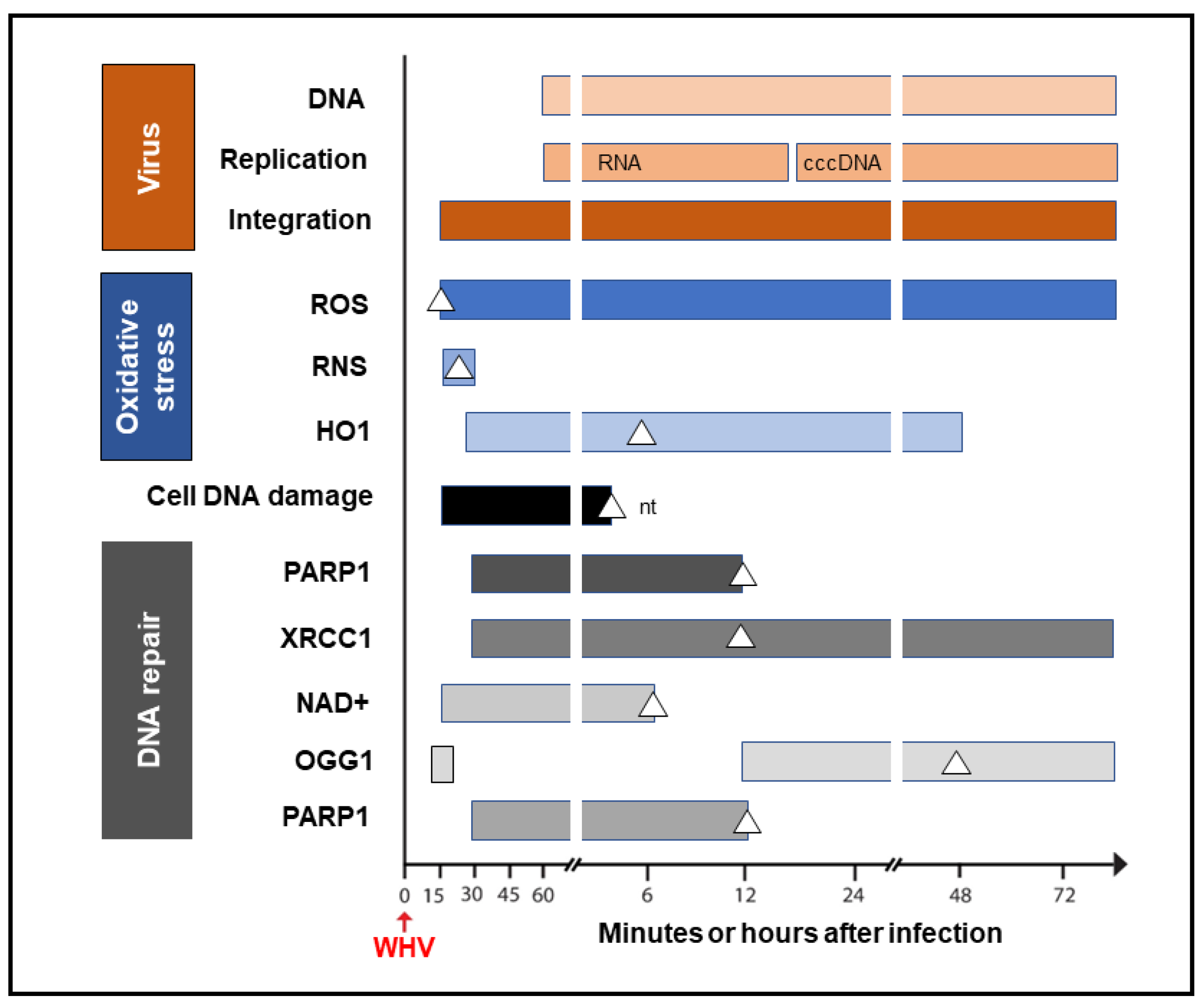
Figure 3. Graphic presentation of the earliest detections and changes over time in the presence of virus genome (DNA), its replication (mRNA and cccDNA) and integration into the hepatocellular genome, indicators of hepatocyte oxidative stress and oxidative DNA damage, as well as markers of the activity of components of the DNA repair pathway during the first 72 h after exposure to HBV or WHV. The profiles represent cumulative data from HBV and WHV infections in human and woodchuck hepatocyte-compatible cells and in woodchucks infected with WHV. White triangles mark the time at which peak expression or activity was found. Whites square in the OGG1 line represent a 5.6-fold increase in the gene expression over uninfected control cells, which did not achieve a statistically significant difference. For more details, see reference [4]. Abbreviations: cccDNA, virus covalently closed circular DNA; ROS, reactive oxygen species; RNS, reactive nitrogen species; HO1, heme oxygenase-1; PARP1, poly(ADP-ribose) polymerase 1; XRCC1, X-ray repair cross-complementing protein 1; NAD+, nicotinamide adenine dinucleotide; OGG1, 8-oxyguanidine DNA glucose 1, and nt, not tested beyond this time point.
In addition, a transient 5.6-fold increase over uninfected control cells in the OGG1 gene expression (although not statistically significant) was detected between 15 and 30 min p.i. and then from 12 h p.i. onward [4]. PARP1 cleavage became significantly augmented between 6 and 12 h p.i., but a twofold increase, although not statistically significant, was identified 1 h after infection. These quantitative measurements indicate that hepadnavirus is a strong and essentially immediate inducer of hepatocyte oxidative stress and associated DNA damage and that the PARP1/XRRC1-initiated dsDNA NHEJ repair is involved in the creation of the earliest virus–host fusions. A progressive increase in PARP1 transcription up to 6 h p.i. subsided after 12 h p.i. to the level detectable in control cells (Figure 3). This sharp decline was accompanied by an increase in the cleavage of PARP1 protein (see Figure 5B presented in reference [4]). Jointly, researchers' data show that PARP1-facilitated dsDNA repair is engaged in the initial stages of infection, and the NHEJ mechanism determines the HTJ format of the earliest hepadnavirus–host genomic merges. In stages beyond a few hours after infection, the virus–host junctions tend to display the OHJ format more often, implying their creation by the MHOJ repair mechanism.
References
- Chauhan, R.; Churchill, N.D.; Mulrooney-Cousins, P.M.; Michalak, T.I. Initial sites of hepadnavirus integration into host genome in human hepatocytes and in the woodchuck model of hepatitis B-associated hepatocellular carcinoma. Oncogenesis 2017, 6, e317.
- Chauhan, R.; Shimizu, Y.; Watashi, K.; Wakita, T.; Fukasawa, M.; Michalak, T.I. Retrotransposon elements among initial sites of hepatitis B virus integration into human genome in the HepG2-NTCP cell infection model. Cancer Genet. 2019, 235–236, 39–56.
- Tu, T.; Budzinska, M.A.; Vondran, F.W.R.; Shackel, N.A.; Urban, S. Hepatitis B Virus DNA Integration Occurs Early in the Viral Life Cycle in an In Vitro Infection Model via Sodium Taurocholate Cotransporting Polypeptide-Dependent Uptake of Enveloped Virus Particles. J. Virol. 2018, 92, e02007-17.
- Chauhan, R.; Michalak, T.I. Kinetics of DNA damage repair response accompanying initial hepadnavirus-host genomic integration in woodchuck hepatitis virus infection of hepatocyte. Cancer Genet. 2020, 244, 1–10.
- Audebert, M.; Salles, B.; Calsou, P. Involvement of Poly(ADP-ribose) Polymerase-1 and XRCC1/DNA Ligase III in an Alternative Route for DNA Double-strand Breaks Rejoining. J. Biol. Chem. 2004, 279, 55117–55126.
- Wang, M.; Wu, W.; Wu, W.; Rosidi, B.; Zhang, L.; Wang, H.; Iliakis, G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006, 34, 6170–6182.
- Mason, W.S.; Gill, U.S.; Litwin, S.; Zhou, Y.; Peri, S.; Pop, O.; Hong, M.L.; Naik, S.; Quaglia, A.; Bertoletti, A.; et al. HBV DNA Integration and Clonal Hepatocyte Expansion in Chronic Hepatitis B Patients Considered Immune Tolerant. Gastroenterology 2016, 151, 986–998.e4.
- Dandri, M.; Burda, M.R.; Bürkle, A.; Zuckerman, D.M.; Will, H.; Rogler, C.E.; Greten, H.; Petersen, J. Increase in de novo HBV DNA integrations in response to oxidative DNA damage or inhibition of poly(ADP-ribosyl)ation. Hepatology 2002, 35, 217–223.
More
