Circular RNA (CircRNA), a single-stranded closed-loop RNA that lacks terminal 5′ caps and 3′ poly (A) tails, is more stable than linear RNA. CircRNA was first discovered in plant-infected viroids and was later observed in eukaryotes, although it was not produced via the back-splicing mechanism.
- circRNA
- OSCC
- biomarker
- miRNA sponge
1. Characteristics
Circular RNA (CircRNA), a single-stranded closed-loop RNA that lacks terminal 5′ caps and 3′ poly (A) tails, is more stable than linear RNA [8][1]. CircRNA was first discovered in plant-infected viroids [9][2] and was later observed in eukaryotes, although it was not produced via the back-splicing mechanism [10][3]. Increasing numbers of differentially expressed circRNAs have been identified as a result of improvements in high-throughput sequencing techniques and bioinformatics. Over the years, a variety of the biological functions of circRNAs have been disclosed. An increasing number of studies have suggested that circRNAs are involved in a variety of diseases and may serve as biomarkers for cancer diagnostic and therapeutic targets [11][4].
-
Diversity: from fruit flies to humans, circRNAs have been discovered in a diverse range of metazoans, cell types, and species. Additionally, circRNAs have been detected in plants and other organisms;
-
Stability: due to their distinctive topology, circRNAs, lacking 5′ caps and 3′ tails in comparison with other non-coding RNAs, are more stable than linear RNAs [13][6]. In addition, owing to their resistance to RNA exonucleases or RNase R [14][7], circRNAs exhibit a half-life over 48 h in most species [13][6];
2. Biogenesis
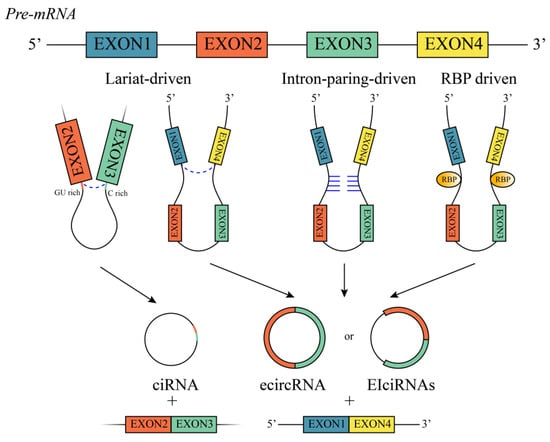
3. Functions
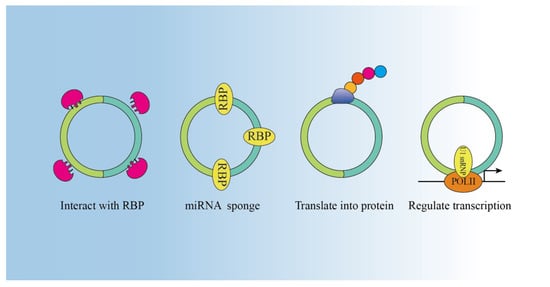
3.1. miRNAs Sponge
3.2. Interaction with RNA Binding Proteins (RBPs)
3.3. Encoded Proteins
3.4. Transcription Regulation
References
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.-M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e13.
- Zhang, Z.; Qi, S.; Tang, N.; Zhang, X.; Chen, S.; Zhu, P.; Ma, L.; Cheng, J.; Xu, Y.; Lu, M.; et al. Discovery of Replicating Circular RNAs by RNA-Seq and Computational Algorithms. PLoS Pathog. 2014, 10, e1004553.
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular Transcripts of the Testis-Determining Gene Sry in Adult Mouse Testis. Cell 1993, 73, 1019–1030.
- Verduci, L.; Tarcitano, E.; Strano, S.; Yarden, Y.; Blandino, G. CircRNAs: Role in Human Diseases and Potential Use as Biomarkers. Cell Death Dis. 2021, 12, 468.
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS ONE 2012, 7, e30733.
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs Are Abundant, Conserved, and Associated with ALU Repeats. RNA 2013, 19, 141–157.
- Suzuki, H.; Zuo, Y.; Wang, J.; Zhang, M.Q.; Malhotra, A.; Mayeda, A. Characterization of RNase R-Digested Cellular RNA Source That Consists of Lariat and Circular RNAs from Pre-mRNA Splicing. Nucleic Acids Res. 2006, 34, e63.
- AbouHaidar, M.G.; Venkataraman, S.; Golshani, A.; Liu, B.; Ahmad, T. Novel Coding, Translation, and Gene Expression of a Replicating Covalently Closed Circular RNA of 220 Nt. Proc. Natl. Acad. Sci. USA 2014, 111, 14542–14547.
- Drula, R.; Braicu, C.; Harangus, A.; Nabavi, S.M.; Trif, M.; Slaby, O.; Ionescu, C.; Irimie, A.; Berindan-Neagoe, I. Critical Function of Circular RNAs in Lung Cancer. Wiley Interdiscip. Rev. RNA 2020, 11, e1592.
- Bach, D.-H.; Lee, S.K.; Sood, A.K. Circular RNAs in Cancer. Mol. Ther. Nucleic Acids 2019, 16, 118–129.
- Zhang, Y.; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular Intronic Long Noncoding RNAs. Mol. Cell 2013, 51, 792–806.
- Hatibaruah, A.; Rahman, M.; Agarwala, S.; Singh, S.A.; Shi, J.; Gupta, S.; Paul, P. Circular RNAs in Cancer and Diabetes. J. Genet. 2021, 100, 21.
- Okholm, T.L.H.; Sathe, S.; Park, S.S.; Kamstrup, A.B.; Rasmussen, A.M.; Shankar, A.; Chua, Z.M.; Fristrup, N.; Nielsen, M.M.; Vang, S.; et al. Transcriptome-Wide Profiles of Circular RNA and RNA-Binding Protein Interactions Reveal Effects on Circular RNA Biogenesis and Cancer Pathway Expression. Genome Med. 2020, 12, 112.
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233.
- Ouyang, Y.; Li, Y.; Huang, Y.; Li, X.; Zhu, Y.; Long, Y.; Wang, Y.; Guo, X.; Gong, K. CircRNA circPDSS1 Promotes the Gastric Cancer Progression by Sponging miR-186-5p and Modulating NEK2. J. Cell Physiol. 2019, 234, 10458–10469.
- Han, D.; Li, J.; Wang, H.; Su, X.; Hou, J.; Gu, Y.; Qian, C.; Lin, Y.; Liu, X.; Huang, M.; et al. Circular RNA circMTO1 Acts as the Sponge of microRNA-9 to Suppress Hepatocellular Carcinoma Progression. Hepatology 2017, 66, 1151–1164.
- Zhou, Y.; Zhang, S.; Min, Z.; Yu, Z.; Zhang, H.; Jiao, J. Knockdown of Circ_0011946 Targets miR-216a-5p/BCL2L2 Axis to Regulate Proliferation, Migration, Invasion and Apoptosis of Oral Squamous Cell Carcinoma Cells. BMC Cancer 2021, 21, 1085.
- Ghafouri-Fard, S.; Khoshbakht, T.; Taheri, M.; Jamali, E. A Concise Review on the Role of CircPVT1 in Tumorigenesis, Drug Sensitivity, and Cancer Prognosis. Front. Oncol. 2021, 11, 762960.
- Goutas, D.; Pergaris, A.; Giaginis, C.; Theocharis, S. HuR as Therapeutic Target in Cancer: What the Future Holds. Curr. Med. Chem. 2022, 29, 56–65.
- Li, K.; Fan, X.; Yan, Z.; Zhan, J.; Cao, F.; Jiang, Y. Circ_0000745 Strengthens the Expression of CCND1 by Functioning as miR-488 Sponge and Interacting with HuR Binding Protein to Facilitate the Development of Oral Squamous Cell Carcinoma. Cancer Cell Int. 2021, 21, 271.
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e7.
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA That Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e9.
- Zhang, M.; Huang, N.; Yang, X.; Luo, J.; Yan, S.; Xiao, F.; Chen, W.; Gao, X.; Zhao, K.; Zhou, H.; et al. A Novel Protein Encoded by the Circular Form of the SHPRH Gene Suppresses Glioma Tumorigenesis. Oncogene 2018, 37, 1805–1814.
- Peng, Y.; Xu, Y.; Zhang, X.; Deng, S.; Yuan, Y.; Luo, X.; Hossain, M.T.; Zhu, X.; Du, K.; Hu, F.; et al. A Novel Protein AXIN1-295aa Encoded by circAXIN1 Activates the Wnt/β-Catenin Signaling Pathway to Promote Gastric Cancer Progression. Mol. Cancer 2021, 20, 158.
- Zlotorynski, E. Non-Coding RNA: Circular RNAs Promote Transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 206.
