Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Camila Xu and Version 1 by Kyoungtae Kim.
Quantum dots (QDs) are a type of nanoparticle with exceptional photobleaching-resistant fluorescence. They are highly sought after for their potential use in various optical-based biomedical applications.
- quantum dots
- biomedical applications
- nanomaterials
1. Introduction
Ever since their discovery in 1981, quantum dots (QDs) have intrigued scientific minds worldwide with their unique physical and optical properties. Quantum dots are nano-sized semiconductor crystals with a broad emission range [1]. Their emission spectra are closely related to their diameter [2,3,4,5,6,7][2][3][4][5][6][7]. This specific characteristic of quantum dots allows for easy manipulation of QDs’ size to achieve the desired fluorescence color during production. Quantum dots are also superior to many other fluorescence probes, such as organic dyes, since QDs’ fluorescence is known to be photobleaching-resistant [8,9,10,11,12][8][9][10][11][12]. Due to their physical properties, the structure of quantum dots is relatively stable. In some recent studies, QDs have been demonstrated to remain intact under various pH levels [13], high UV exposure [12], and exposure to oxidative conditions [14]. A standard structure of QDs is composed of a core—often heavy metals—encapsulated by a protective shell [15,16][15][16]. QDs can also be conjugated with different types of ligands, which often dictate QDs’ interaction with their surrounding environment [17,18,19,20,21][17][18][19][20][21]. The addition of a protective shell and surface ligands increases the solubility and strengthens the structure of QDs [9,11,15,22,23,24][9][11][15][22][23][24]. The protective shell also greatly reduces the exposure of the heavy metal contents residing in the core of QDs [25[25][26],26], considered to be one of the main mechanisms of QDs’ toxicity. Another characteristic of quantum dots is that they can be synthesized using many methods. However, these methods are often categorized into either top-down or bottom-up approaches [27]. In the top-down approach, larger precursors are used to form smaller QDs as products through hydrothermal methods [28], electrochemical methods [29], laser ablation [30], etc. On the other hand, the bottom-up approach uses smaller precursors, such as organic molecules, to build larger QD structures through pyrolysis [31] and heat [32]. Although each approach has its own advantages and disadvantages [33], the variety of methods enables developers to choose the best-suited approach.
Due to the diverse types of available quantum dots, QDs are often categorized by their core composition. One of the most widely used cores for quantum dots is cadmium. Cadmium-based QDs, such as cadmium selenide (CdSe), cadmium telluride (CdTe), and cadmium sulfide (CdS), are of great interest to researchers due to their phenomenally high quantum yield compared to other types of QDs [34,35,36][34][35][36]. Thus, cadmium QDs are an excellent candidate for biomedical applications such as trackable drug delivery and bioimaging. However, the potential for cadmium-based QD usage for these biological purposes is currently hindered due to the potential toxicity of their cadmium core [37,38,39,40,41][37][38][39][40][41]. Cadmium is well known to cause many major problems for the environment, as well as for human health [42,43,44,45][42][43][44][45]. It has been reported that cadmium exposure can lead to serious diseases in adults [46] and cause detrimental developmental impairments in children [47]. The United States Environmental Protection Agency (EPA) has also classified cadmium as a probable carcinogenic agent for humans [48]. As such, the usage of cadmium-based QDs raises concerns for many people due to the possible leakage of cadmium ions. In response, countless studies have been dedicated to studying the impact of QDs on the cellular, molecular, tissue, and even organismal levels [49,50,51,52,53,54,55,56][49][50][51][52][53][54][55][56]. In the past decade, many studies have reported the toxicity of cadmium-based QDs, including inducing ROS levels [51[51][57],57], triggering apoptosis [57], alternate gene expression profiles [58,59[58][59][60],60], damaging the structure and function of mitochondria [61], negative impact on the reproductive system [62[62][63],63], causing neurotoxicity [64], and many other undesirable side effects. Due to the countless negative impacts, cadmium QDs are often deemed to be unideal for in vivo biomedical applications. Thus, researchers have shifted their attention to developing other types of QDs that do not contain cadmium.
Indium-based QDs were among the earliest developed non-cadmium QDs. The recent literature suggests that indium-based QDs are less toxic compared to cadmium QDs. Several studies have found that compared to cadmium-based QDs, indium-based QDs have less impact on cell viability, induce less DNA damage, and are overall not toxic [65]. However, contradictory results have also been reported. Davenport et al., reported a similar toxic effect on cell viability between cadmium selenide/zinc sulfide quantum dots (CdSe/ZnS QDs) and indium phosphide/zinc sulfide quantum dots (InP/ZnS QDs) [66]. Cullen et al., found that InP/ZnS QDs reduced the endpoint optical density of the budding yeast Saccharomyces cerevisiae, while CdSe/ZnS QDs only caused a prolonged lag phase but did not greatly impact the final optical density [58]. Therefore, more studies need to be conducted before indium-based QDs could be used as a safe alternative. In addition to indium-based QDs, other types of QDs such as copper-based QDs [67], silver-based QDs [68], and many others have also been developed. However, these quantum dots have also been shown to have a negative impact on cells. For example, with ternary copper indium disulfide/zinc sulfide quantum dots (CulnS2/ZnS QDs), a high dose induced a minor inflammatory response in the lymph nodes of mice [69]. Silver quantum dots have been reported to exert toxicity by interfering with plants’ photosynthesis processes [70]. Thus, collectively, these studies suggest that each type of QDs has its own toxicity mechanism that needs to be further investigated. Another alternative to cadmium quantum dots is doped quantum dots. Among these, manganese-doped zinc sulfide dots (Mn: ZnS d-dots) are considered to have the most potential due to their low toxicity [71]. Furthermore, Mn-doped QDs have been shown to possess both fluorescence and magnetic properties, making them an ideal candidate for multimodal imaging [72].
2. Ligands
In addition to using alternative core types, modifications to other QD components, such as the surface ligand, have also been explored to maximize QDs’ biocompatibility and efficacy for in vivo applications. Surface ligands are essential for the interaction between QDs and the surrounding environment [17]. Therefore, understanding the behavior of QDs with different conjugated ligands is the first step toward picking the right ligand type for potential clinical usage. In the pharmaceutical field, one of the key factors for an agent to be considered for biological applications is solubility [75][73]. Thus, numerous strategies have been developed to improve the solubility of chemical treatments [76,77,78][74][75][76] and drug delivery carriers [79,80,81][77][78][79]. For QDs, solubility issues can be avoided by coating the nanocrystal with hydrophilic surface ligands. For example, Ghani et al., replaced the original hydrophobic tri-octyl phosphine oxide (TOPO) ligands on CdSe/ZnS QDs with different types of water-soluble bisphosphonate (BIP)-based ligands, such as ethylene diphosphonate (EDP), imido diphosphonate (IDP), and methylene diphosphonate (MDP). The results showed that the exchanged ligand improved the water solubility and dispersal of CdSe/ZnS QDs [22]. Furthermore, EDP-, IDP-, and MDP-conjugated CdSe/ZnS QDs were shown to have lower toxicity effects compared to CdSe/ZnS QDs conjugated with TOPO ligands. Additionally, the study found that EDP and MDP were significantly taken up by IGROV-1 ovarian cancer cells compared to IDP-conjugated CdSe/ZnS QDs [22]. The difference in the cellular uptake of QDs with different target ligands showed that certain ligands are more suitable than others for biological applications. It is also worth noting that the ligand-exchange step could be skipped if QDs are synthesized in an aqueous phase [71], making it easier to conjugate the desired ligands on QDs. In another study, Al-Hajaj et al., found that equally sized CdSe(CdZnS) QDs with different conjugated ligands had distinct modes of entry and were taken up in different quantities. Their data revealed that negatively charged CdSe(CdZnS) QDs-CA (QDs conjugated with cysteamine ligands) entered HEK293 human kidney cells and HepG2 human liver cells at a much higher levels compared to positively charged CdSe(CdZnS) QDs, such as CdSe(CdZnS) QDs-CYS (QDs conjugated with cysteine), CdSe(CdZnS) QDs-MPA (mercaptopropionic acid), and CdSe(CdZnS) QDs-DHLA (dihydrolipoic acid) [21]. Ligand types also had an impact on the rate of CdSe(CdZnS) QDs elimination. CdSe(CdZnS) QDs-CA clearance from HepG2 and HEK293 cells took place in the first 3 h post-treatment, while around 80% of CdSe(CdZnS) QDs-CYS was still retained in HepG2 cells after 6 h post-treatment [21]. In addition to affecting the rate of QDs’ cellular uptake, surface ligand types also influence the interaction of QDs with biological components. In 2023, Yu et al., investigated the interaction and impact of two different ligands on CdSe/ZnS QDs—glutathione (GSH-QDs) and dihydrolipoic acid (DHLA QDs)—on the alpha chymotrypsin (ChT) enzyme. Their data revealed that GSH-QDs weakly inhibited ChT’s catalytic activity, while DHLA-QDs greatly inhibited ChT activity. Both DHLA-QDs and GSH-QDs were able to bind to ChT at a 1-to-8 ratio. However, DHLA-QDs have a greater affinity for ChT compared to GSH-QDs, suggesting that the binding affinity of DHLA-QDs with ChT is one of the key factors for the inhibition of ChT activity. The different ligands also resulted in different binding mechanisms between the QDs and ChT, as DHLA-QDs bound to ChT via hydrophobic interactions, while GSH-QDs bound to ChT through hydrogen bonding and van der Waals forces [20]. Recently, some studies have reported that various types of QDs could also interact with common proteins, such as bovine serum albumin (BSA) [82,83,84][80][81][82]. It has been shown that in the presence of CdSe QDs, BSA could bind and form corona protein complexes [85][83]. The formation of corona protein complexes in the serum could lead to several issues, such as protein structure alteration, protein aggregation, and protein denaturation. In humans, a similar protein called human serum albumin, which is abundant in human serum, has also been reported to form bind to CdSe/ZnS QDs [86][84]. Thus, it is possible that the same phenomenon could also take place when QDs are used for in vivo applications in humans. The interaction between QDs with different ligands and other biological materials is consistent with prior reports, where ligands played an essential role in the binding of proteins to nanoclusters [87][85]. Thus, future research should investigate ligand-dependent interactions between QDs and proteins to develop a modified ligand with minimal unwanted QDs–protein interactions. Collectively, the studies above highlight the importance of choosing the right surface ligands for QDs. Depending on the aim of the application, ligand choice can enhance the safety and efficacy of QDs by altering the interaction between QDs and biological components.3. QDs as a Labeling Agent
Due to their unique characteristics, QDs are vastly useful as a fluorescence label. Previously, Q-tracker 565 and Q-tracker 655 from the Quantum Dots Corporation (Hayward, CA, USA) were shown to effectively label several hematological cell lines (KG-1, HL-60, and SUDHL-16), as well as cells derived from the bone marrow and umbilical cords, by residing in the intracellular space of cells. The labeling of these cells was shown to last from one to two weeks post-incubation. Furthermore, these QDs were found to remain in cells through four cell division cycles, with decreasing QD fluorescence after each cycle. In cells such as HL-60 cells and umbilical-cord-derived CD34+ cells, QDs were also shown to be retained through cell differentiation [88][86]. These findings showed that QDs could reside intracellularly and label different hematological cell lines for an extended period of time, providing evidence that QDs could be a useful tool for hematological cell imaging. However, the same study also revealed that intracellular labeling by QDs was seen in all tested cell lines, hinting that the labeling by these QDs is not selective. As such, the same group of researchers attempted to target QDs to specific cells by conjugating QDs with streptavidin (QDs-SA). To target cells that specifically express CD33 on the cell surface, QDs-SA were incubated with biotinylated anti-CD33 antibodies prior to cell treatment. The results showed that QDs-SA incubated with biotinylated anti-CD33 selectively bind to cells that express CD33 (HL-60) [88][86]. These results indicate that when QDs are not conjugated with a selective ligand, they can be randomly internalized by multiple hematological cell lines, thus providing evidence that the presence of targeting ligands is essential for the specific binding. Apart from whole-cell labeling, QDs have also been used to label specific organelles’ structures. Traditionally, organic dyes are commonly used as fluoroprobes to visualize different structures of cells. However, problems such as short lifetime and weak signal intensity limit their efficiency in cell imaging. As QDs are well known to have photobleaching-resistant fluorescence, they could also be used as superior labeling agents. In one study, streptavidin-conjugated CdSe/ZnS QDs were used to label the actin cytoskeleton of SK-BR-3 cancer cells. The authors found that QDs-streptavidin were able to clearly label the biotinylated F-actin structure of cancer cells. Furthermore, CdSe/ZnS QDs-streptavidin were also able to label other cell structures, such as the nucleus of SK-BR-3 cancer cells pre-incubated with nuclear antigens and biotinylated anti-human IgG [89][87]. In the most recent 2023 study, neutravidin-conjugated CuInS2/ZnS (CIS/ZnS) QDs and neutravidin-conjugated CdSe/ZnS QDs were used as F-actin labeling agents for super-resolution imaging. Although improvements in labeling density are still needed, both QD types effectively labeled F-actin and significantly improved the resolution compared to conventional fluorescence imaging [90][88]. In addition to fixed structural imaging, QDs could also be used as probes to study real-time cellular processes. Hatakeyama et al., used Qdot (QD655) in combination with HaloTag technology to study the dynamics of the cytosolic myosin motor protein. In this study, QDs were conjugated with a HaloTag ligand and electroporated into cells, where HaloTag ligand-QDs found the protein of interest (myosin) that was fused with the HaloTag protein (Figure 1). In this way, QDs were able to indirectly bind to myosin and act as a probe to study its intracellular movements and interactions. Using this technology, the authors were able to observe myosin’s movement along the actin filament [91][89]. In a more recent study by Zhang et al., the combination of QDs and HaloTag proteins was used in a similar manner to study the movement of proteins selectively expressed on the surface of mammalian cells through mammalian display technology. In this case, the HaloTag protein was chosen as the displayed surface protein. QDs conjugated with HaloTag ligands (HTL-QDs) were added to act as a probe to track the movement of the displayed HaloTag protein throughout the experiment. Around 30 min after the temperature shift, the displayed HaloTag protein tagged with HTL-QDs moved from the membrane to the cytoplasm. This indicates that the membrane-displayed protein is not always on the cell membrane but is capable of reentering the cells upon temperature changes. The internalized HLT-QDs were transported by membrane-bound vehicles and located near the nucleus but never entered it. The HLT-QDs signals were eventually either detected in the lysosome or recycled back to the membrane surface [92][90]. Thus, quantum dots are an excellent fluorescence probe for the labeling and imaging of cellular processes.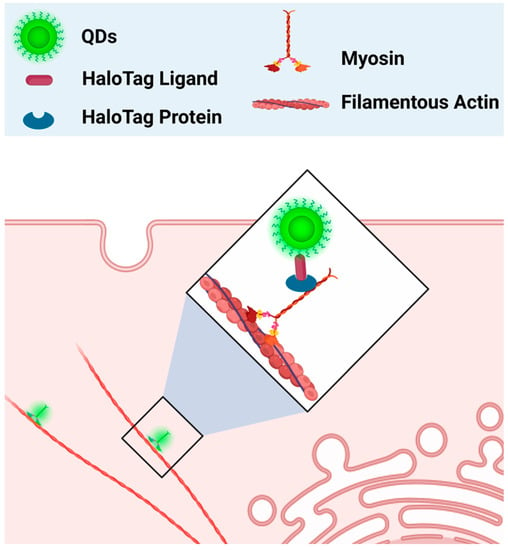
Figure 1. QDs as a fluorescence probe to study the dynamics of myosin’s movement. The HaloTag ligand-QDs complex (HaloTag-QDs) bound to a HaloTag protein that was previously fused with myosin. QDs’ fluorescence helped track the movement of myosin. Diagram created based on findings by Hatakeyama et al. [91][89].
References
- Joglekar, P.; Mandalkar, D.; Nikam, M.; Pande, N.; Dubal, A. Review Article on Quantum Dots: Synthesis, Properties and Application. Int. J. Res. Advent Technol. 2019, 7, 510–515.
- Bhandari, S.; Hao, B.; Waters, K.; Lee, C.H.; Idrobo, J.C.; Zhang, D.; Pandey, R.; Yap, Y.K. Two-Dimensional Gold Quantum Dots with Tunable Bandgaps. ACS Nano 2019, 13, 4347–4353.
- Mirhosseini Moghaddam, M.; Baghbanzadeh, M.; Sadeghpour, A.; Glatter, O.; Kappe, C.O. Continuous-Flow Synthesis of CdSe Quantum Dots: A Size-Tunable and Scalable Approach. Chem.-A Eur. J. 2013, 19, 11629–11636.
- Liu, Z.; Li, F.; Luo, Y.; Li, M.; Hu, G.; Pu, X.; Tang, T.; Wen, J.; Li, X.; Li, W. Size Effect of Graphene Quantum Dots on Photoluminescence. Molecules 2021, 26, 3922.
- Imran, A.; Jiang, J.; Eric, D.; Yousaf, M. Size and Shape Dependent Optical Properties of InAs Quantum Dots. In Proceedings of the 2017 International Conference on Optical Instruments and Technology: Micro/Nano Photonics: Materials and Devices, Beijing, China, 28–30 October 2017; SPIE: Bellingham, WA, USA, 2018; Volume 10622, pp. 54–62.
- Chua, S.J.; Ngo, S.; Yoon, Y.C.; Fan, S.F.; Chua, W.J. Effects of Size and Shape on Electronic States of Quantum Dots. Phys. Rev. B 2006, 74, 245331–245332.
- Liu, Y.; Bose, S.; Fan, W. Effect of Size and Shape on Electronic and Optical Properties of CdSe Quantum Dots. Optik 2018, 155, 242–250.
- Valizadeh, A.; Mikaeili, H.; Samiei, M.; Farkhani, S.M.; Zarghami, N.; Kouhi, M.; Akbarzadeh, A.; Davaran, S. Quantum Dots: Synthesis, Bioapplications, and Toxicity. Nanoscale Res. Lett. 2012, 7, 480.
- Yu, S.; Zhang, X.; Li, L.; Xu, J.; Song, Y.; Liu, X.; Wu, S.; Zhang, J. High Photostability and Luminescent Efficiency of Quantum Dots: Ultrathin Epitaxial Al Self-Passivation Layer with a Homogeneous Ligand. Mater. Res. Express 2019, 6, 0850f7.
- Selopal, G.S.; Zhao, H.; Wang, Z.M.; Rosei, F. Core/Shell Quantum Dots Solar Cells. Adv. Funct. Mater. 2020, 30, 1908762.
- Tan, Y.; Jin, S.; Hamers, R.J. Photostability of Cdse Quantum Dots Functionalized with Aromatic Dithiocarbamate Ligands. ACS Appl. Mater. Interfaces 2013, 5, 12975–12983.
- Bailes, J. Photostability of Semiconductor Quantum Dots in Response to UV Exposure. Methods Mol. Biol. 2020, 2118, 343–349.
- Le, N.; Routh, J.; Kirk, C.; Wu, Q.; Patel, R.; Keyes, C.; Kim, K. Red CdSe/ZnS QDs’ Intracellular Trafficking and Its Impact on Yeast Polarization and Actin Filament. Cells 2023, 12, 484.
- Yeh, C.W.; Chen, G.H.; Ho, S.J.; Chen, H.S. Inhibiting the Surface Oxidation of Low-Cadmim-Content ZnS:(Cd,Se) Quantum Dots for Enhancing Application Reliability. ACS Appl. Nano Mater. 2019, 2, 5290–5301.
- Vasudevan, D.; Gaddam, R.R.; Trinchi, A.; Cole, I. Core-Shell Quantum Dots: Properties and Applications. J. Alloys Compd. 2015, 636, 395–404.
- Wang, Z.; Tang, M. The Cytotoxicity of Core-Shell or Non-Shell Structure Quantum Dots and Reflection on Environmental Friendly: A Review. Environ. Res. 2021, 194, 110593.
- Tekle, C.; Van Deurs, B.; Sandvig, K.; Iversen, T.G. Cellular Trafficking of Quantum Dot-Ligand Bioconjugates and Their Induction of Changes in Normal Routing of Unconjugated Ligands. Nano Lett. 2008, 8, 1858–1865.
- Green, M. The Nature of Quantum Dot Capping Ligands. J. Mater. Chem. 2010, 20, 5797–5809.
- Tan, S.J.; Jana, N.R.; Gao, S.; Patra, P.K.; Ying, J.Y. Surface-Ligand-Dependent Cellular Interaction, Subcellular Localization, and Cytotoxicity of Polymer-Coated Quantum Dots. Chem. Mater. 2010, 22, 2239–2247.
- Yu, Y.Q.; Chen, W.Q.; Li, X.H.; Liu, M.; He, X.H.; Liu, Y.; Jiang, F.L. Quantum Dots Meet Enzymes: Hydrophobicity of Surface Ligands and Size Do Matter. Langmuir 2023, 39, 3967–3978.
- Al-Hajaj, N.A.; Moquin, A.; Neibert, K.D.; Soliman, G.M.; Winnik, F.M.; Maysinger, D. Short Ligands Affect Modes of QD Uptake and Elimination in Human Cells. ACS Nano 2011, 5, 4909–4918.
- Abdul Ghani, S.F.; Wright, M.; Paramo, J.G.; Bottrill, M.; Green, M.; Long, N.; Thanou, M. Three Bisphosphonate Ligands Improve the Water Solubility of Quantum Dots. Faraday Discuss. 2014, 175, 153–169.
- Zhang, Y.; Schnoes, A.M.; Clapp, A.R. Dithiocarbamates as Capping Ligands for Water-Soluble Quantum Dots. ACS Appl. Mater. Interfaces 2010, 2, 3384–3395.
- Nam, E.; Lee, C.; Kim, S.J.; Chung, H.K.; Chae, H. Stability and Dispersion Improvement of Quantum-Dot Films by Hydrosilylation between Quantum-Dot Ligands and a Siloxane Matrix. Opt. Express 2019, 27, 20037–20046.
- Mei, J.; Yang, L.Y.; Lai, L.; Xu, Z.Q.; Wang, C.; Zhao, J.; Jin, J.C.; Jiang, F.L.; Liu, Y. The Interactions between CdSe Quantum Dots and Yeast Saccharomyces Cerevisiae: Adhesion of Quantum Dots to the Cell Surface and the Protection Effect of ZnS Shell. Chemosphere 2014, 112, 92–99.
- Wu, T.; Liang, X.; He, K.; Liu, X.; Li, Y.; Wang, Y.; Kong, L.; Tang, M. The NLRP3-Mediated Neuroinflammatory Responses to Cdte Quantum Dots and the Protection of ZnS Shell. Int. J. Nanomed. 2020, 15, 3217–3233.
- Nocito, G.; Calabrese, G.; Forte, S.; Petralia, S.; Puglisi, C.; Campolo, M.; Esposito, E.; Conoci, S. Carbon Dots as Promising Tools for Cancer Diagnosis and Therapy. Cancers 2021, 13, 1991.
- Wang, K.; Dong, J.; Sun, L.; Chen, H.; Wang, Y.; Wang, C.; Dong, L. Effects of Elemental Doping on the Photoluminescence Properties of Graphene Quantum Dots. RSC Adv. 2016, 6, 91225–91232.
- Li, Y.; Li, S.; Wang, Y.; Wang, J.; Liu, H.; Liu, X.; Wang, L.; Liu, X.; Xue, W.; Ma, N. Electrochemical Synthesis of Phosphorus-Doped Graphene Quantum Dots for Free Radical Scavenging. Phys. Chem. Chem. Phys. 2017, 19, 11631–11638.
- Kaczmarek, A.; Hoffman, J.; Morgiel, J.; Mościcki, T.; Stobiński, L.; Szymański, Z.; Małolepszy, A. Luminescent Carbon Dots Synthesized by the Laser Ablation of Graphite in Polyethylenimine and Ethylenediamine. Materials 2021, 14, 729.
- Lotz, K.; Wütscher, A.; Düdder, H.; Berger, C.M.; Russo, C.; Mukherjee, K.; Schwaab, G.; Havenith, M.; Muhler, M. Tuning the Properties of Iron-Doped Porous Graphitic Carbon Synthesized by Hydrothermal Carbonization of Cellulose and Subsequent Pyrolysis. ACS Omega 2019, 4, 4448–4460.
- Luo, H.; Kebede, B.A.; McLaurin, E.J.; Chikan, V. Rapid Induction and Microwave Heat-Up Syntheses of CdSe Quantum Dots. ACS Omega 2018, 3, 5399–5405.
- Bruno, F.; Sciortino, A.; Buscarino, G.; Soriano, M.L.; Ríos, Á.; Cannas, M.; Gelardi, F.; Messina, F.; Agnello, S. A Comparative Study of Top-Down and Bottom-Up Carbon Nanodots and Their Interaction with Mercury Ions. Nanomaterials 2021, 11, 1265.
- Zhimin, Y.; Wang, J.; Yang, P. Highly Luminescent CdTe/CdS/ZnO Core/Shell/Shell Quantum Dots Fabricated Using an Aqueous Strategy. Luminescence 2013, 28, 169–175.
- Kloepfer, J.A.; Bradforth, S.E.; Nadeau, J.L. Photophysical Properties of Biologically Compatible CdSe Quantum Dot Structures. J. Phys. Chem. B 2005, 109, 9996–10003.
- Borovaya, M.N.; Burlaka, O.M.; Naumenko, A.P.; Blume, Y.B.; Yemets, A.I. Extracellular Synthesis of Luminescent CdS Quantum Dots Using Plant Cell Culture. Nanoscale Res. Lett. 2016, 11, 100.
- Chen, N.; He, Y.; Su, Y.; Li, X.; Huang, Q.; Wang, H.; Zhang, X.; Tai, R.; Fan, C. The Cytotoxicity of Cadmium-Based Quantum Dots. Biomaterials 2012, 33, 1238–1244.
- Mo, D.; Hu, L.; Zeng, G.; Chen, G.; Wan, J.; Yu, Z.; Huang, Z.; He, K.; Zhang, C.; Cheng, M. Cadmium-Containing Quantum Dots: Properties, Applications, and Toxicity. Appl. Microbiol. Biotechnol. 2017, 101, 2713–2733.
- Luo, Y.H.; Wu, S.B.; Wei, Y.H.; Chen, Y.C.; Tsai, M.H.; Ho, C.C.; Lin, S.Y.; Yang, C.S.; Lin, P. Cadmium-Based Quantum Dot Induced Autophagy Formation for Cell Survival via Oxidative Stress. Chem. Res. Toxicol. 2013, 26, 662–673.
- Rzigalinski, B.A.; Strobl, J.S. Cadmium-Containing Nanoparticles: Perspectives on Pharmacology and Toxicology of Quantum Dots. Toxicol. Appl. Pharmacol. 2009, 238, 280–288.
- Bechu, A.; Liao, J.; Huang, C.; Ahn, C.; McKeague, M.; Ghoshal, S.; Moores, A. Cadmium-Containing Quantum Dots Used in Electronic Displays: Implications for Toxicity and Environmental Transformations. ACS Appl. Nano Mater. 2021, 4, 8417–8428.
- Branca, J.J.V.; Morucci, G.; Pacini, A. Cadmium-Induced Neurotoxicity: Still Much Ado. Neural Regen. Res. 2018, 13, 1879–1882.
- Haider, F.U.; Liqun, C.; Coulter, J.A.; Cheema, S.A.; Wu, J.; Zhang, R.; Wenjun, M.; Farooq, M. Cadmium Toxicity in Plants: Impacts and Remediation Strategies. Ecotoxicol. Environ. Saf. 2021, 211, 111887.
- Jacobson, T.; Priya, S.; Sharma, S.K.; Andersson, S.; Jakobsson, S.; Tanghe, R.; Ashouri, A.; Rauch, S.; Goloubinoff, P.; Christen, P.; et al. Cadmium Causes Misfolding and Aggregation of Cytosolic Proteins in Yeast. Mol. Cell. Biol. 2017, 37, e00490-16.
- Marmiroli, M.; Mussi, F.; Pagano, L.; Imperiale, D.; Lencioni, G.; Villani, M.; Zappettini, A.; White, J.C.; Marmiroli, N. Cadmium Sulfide Quantum Dots Impact Arabidopsis Thaliana Physiology and Morphology. Chemosphere 2020, 240, 124856.
- Godt, J.; Scheidig, F.; Grosse-Siestrup, C.; Esche, V.; Brandenburg, P.; Reich, A.; Groneberg, D.A. The Toxicity of Cadmium and Resulting Hazards for Human Health. J. Occup. Med. Toxicol. 2006, 1, 22.
- Chandravanshi, L.; Shiv, K.; Kumar, S. Developmental Toxicity of Cadmium in Infants and Children: A Review. Environ. Anal. Health Toxicol. 2021, 36, e2021003.
- Cadmium Compounds (A) Hazard Summary. Available online: https://www.epa.gov/sites/default/files/2016-09/documents/cadmium-compounds.pdf (accessed on 7 May 2023).
- Liang, Y.; Zhang, T.; Tang, M. Toxicity of Quantum Dots on Target Organs and Immune System. J. Appl. Toxicol. 2022, 42, 17–40.
- Duan, J.; Yu, Y.; Li, Y.; Yu, Y.; Li, Y.; Huang, P.; Zhou, X.; Peng, S.; Sun, Z. Developmental Toxicity of CdTe QDs in Zebrafish Embryos and Larvae. J. Nanoparticle Res. 2013, 15, 1700.
- Zhang, T.; Hu, Y.; Tang, M.; Kong, L.; Ying, J.; Wu, T.; Xue, Y.; Pu, Y. Liver Toxicity of Cadmium Telluride Quantum Dots (CdTe QDs) Due to Oxidative Stress in Vitro and in Vivo. Int. J. Mol. Sci. 2015, 16, 23279–23299.
- Chen, Y.; Yang, Y.; Ou, F.; Liu, L.; Liu, X.-H.; Wang, Z.-J.; Jin, L. InP/ZnS QDs Exposure Induces Developmental Toxicity in Rare Minnow (Gobiocypris Rarus) Embryos. Environ. Toxicol. Pharmacol. 2018, 60, 28–36.
- Zheng, N.; Yan, J.; Qian, W.; Song, C.; Zuo, Z.; He, C. Comparison of Developmental Toxicity of Different Surface Modified CdSe/ZnS QDs in Zebrafish Embryos. J. Environ. Sci. 2021, 100, 240–249.
- Zhao, L.; Zong, W.; Zhang, H.; Liu, R. Kidney Toxicity and Response of Selenium Containing Protein-Glutathione Peroxidase (Gpx3) to CdTe QDs on Different Levels. Toxicol. Sci. 2019, 168, 201–208.
- Le, N.; Zhang, M.; Kim, K. Quantum Dots and Their Interaction with Biological Systems. Int. J. Mol. Sci. 2022, 23, 10763.
- Hens, B.; Smothers, J.; Rizvanovic, H.; Patel, R.; Wu, Q.; Kim, K. The Future of Anticancer Drugs: A Cytotoxicity Assessment Study of CdSe/ZnS Quantum Dots. J. Nanotheranostics 2020, 1, 19–38.
- Nguyen, K.C.; Willmore, W.G.; Tayabali, A.F. Cadmium Telluride Quantum Dots Cause Oxidative Stress Leading to Extrinsic and Intrinsic Apoptosis in Hepatocellular Carcinoma HepG2 Cells. Toxicology 2013, 306, 114–123.
- Horstmann, C.; Kim, K. Comparing Transcriptome Profiles of Saccharomyces Cerevisiae Cells Exposed to Cadmium Selenide/Zinc Sulfide and Indium Phosphide/Zinc Sulfide. Genes 2021, 12, 428.
- Horstmann, C.; Davenport, V.; Zhang, M.; Peters, A.; Kim, K. Transcriptome Profile Alterations with Carbon Nanotubes, Quantum Dots, and Silver Nanoparticles: A Review. Genes 2021, 12, 794.
- Horstmann, C.; Kim, D.S.; Campbell, C.; Kim, K. Transcriptome Profile Alteration with Cadmium Selenide/Zinc Sulfide Quantum Dots in Saccharomyces Cerevisiae. Biomolecules 2019, 9, 653.
- Nguyen, K.C.; Rippstein, P.; Tayabali, A.F.; Willmore, W.G. Mitochondrial Toxicity of Cadmium Telluride Quantum Dot Nanoparticles in Mammalian Hepatocytes. Toxicol. Sci. 2015, 146, 31–42.
- Xu, G.; Lin, S.; Law, W.C.; Roy, I.; Lin, X.; Mei, S.; Ma, H.; Chen, S.; Niu, H.; Wang, X. The Invasion and Reproductive Toxicity of QDs-Transferrin Bioconjugates on Preantral Follicle in Vitro. Theranostics 2012, 2, 734–745.
- Yan, S.-Q.; Xing, R.; Zhou, Y.-F.; Li, K.-L.; Su, Y.-Y.; Qiu, J.-F.; Zhang, Y.-H.; Zhang, K.-Q.; He, Y.; Lu, X.-P.; et al. Reproductive Toxicity and Gender Differences Induced by Cadmium Telluride Quantum Dots in an Invertebrate Model Organism. Sci. Rep. 2016, 6, srep34182.
- Tang, M.; Xing, T.; Zeng, J.; Wang, H.; Li, C.; Yin, S.; Yan, D.; Deng, H.; Liu, J.; Wang, M.; et al. Unmodified CdSe Quantum Dots Induce Elevation of Cytoplasmic Calcium Levels and Impairment of Functional Properties of Sodium Channels in Rat Primary Cultured Hippocampal Neurons. Environ. Health Perspect. 2008, 116, 915–922.
- Yaghini, E.; Turner, H.; Pilling, A.; Naasani, I.; MacRobert, A.J. In Vivo Biodistribution and Toxicology Studies of Cadmium-Free Indium-Based Quantum Dot Nanoparticles in a Rat Model. Nanomedicine 2018, 14, 2644–2655.
- Davenport, V. An Assessment of InP/ZnS as Potential Anti-cancer Therapy: Quantum Dot Treatment Induces Stress on HeLa Cells. FASEB J. 2021, 35, 16–32.
- He, L.; Yang, L.; Liu, B.; Zhang, J.; Zhang, C.; Liu, S.; Chen, S.; Zapien, J.A.; Alamry, K.A.; Asiri, A.M.; et al. One-Pot Synthesis of Color-Tunable Copper Doped Zinc Sulfide Quantum Dots for Solid-State Lighting Devices. J. Alloys Compd. 2019, 787, 537–542.
- Hunt, N.J.; Lockwood, G.P.; Le Couteur, F.H.; McCourt, P.A.G.; Singla, N.; Kang, S.W.S.; Burgess, A.; Kuncic, Z.; Le Couteur, D.G.; Cogger, V.C. Rapid Intestinal Uptake and Targeted Delivery to the Liver Endothelium Using Orally Administered Silver Sulfide Quantum Dots. ACS Nano 2020, 14, 1492–1507.
- Pons, T.; Pic, E.; Lequeux, N.; Cassette, E.; Bezdetnaya, L.; Guillemin, F.; Marchal, F.; Dubertret, B. Cadmium-Free CuInS2/ZnS Quantum Dots for Sentinel Lymph Node Imaging with Reduced Toxicity. ACS Nano 2010, 4, 2531–2538.
- Subalya, M.; Voleti, R.; Wait, D.A. The Effects of Different Solid Content Carbon Nanotubes and Silver Quantum Dots on Potential Toxicity to Plants through Direct Effects on Carbon and Light Reactions of Photosynthesis. WSEAS Trans. Electron. 2022, 13, 11–18.
- Geszke-Moritz, M.; Piotrowska, H.; Murias, M.; Balan, L.; Moritz, M.; Lulek, J.; Schneider, R. Thioglycerol-Capped Mn-Doped ZnS Quantum Dot Bioconjugates as Efficient Two-Photon Fluorescent Nano-Probes for Bioimaging. J. Mater. Chem. B 2013, 1, 698–706.
- Zhou, R.; Sun, S.; Li, C.; Wu, L.; Hou, X.; Wu, P. Enriching Mn-Doped ZnSe Quantum Dots onto Mesoporous Silica Nanoparticles for Enhanced Fluorescence/Magnetic Resonance Imaging Dual-Modal Bio-Imaging. ACS Appl. Mater. Interfaces 2018, 10, 34060–34067.
- Khan, K.U.; Minhas, M.U.; Badshah, S.F.; Suhail, M.; Ahmad, A.; Ijaz, S. Overview of Nanoparticulate Strategies for Solubility Enhancement of Poorly Soluble Drugs. Life Sci. 2022, 291, 120301.
- Xu, Z.; Lin, S.; Li, Q.; Jiang, S.; Wang, P. Recent Advances in Techniques for Enhancing the Solubility of Hydrophobic Drugs. Pak. J. Pharm. Sci. 2022, 35, 95–113.
- Tran, P.H.L.; Duan, W.; Lee, B.J.; Tran, T.T.D. The Use of Zein in the Controlled Release of Poorly Water-Soluble Drugs. Int. J. Pharm. 2019, 566, 557–564.
- Barrett, J.A.; Yang, W.; Skolnik, S.M.; Belliveau, L.M.; Patros, K.M. Discovery Solubility Measurement and Assessment of Small Molecules with Drug Development in Mind. Drug Discov. Today 2022, 27, 1315–1325.
- Göke, K.; Bunjes, H. Drug Solubility in Lipid Nanocarriers: Influence of Lipid Matrix and Available Interfacial Area. Int. J. Pharm. 2017, 529, 617–628.
- Kumar, S.; Bhargava, D.; Thakkar, A.; Arora, S. Drug Carrier Systems for Solubility Enhancement of BCS Class II Drugs: A Critical Review. Crit. Rev. Ther. Drug Carr. Syst. 2013, 30, 217–256.
- Ghezzi, M.; Pescina, S.; Padula, C.; Santi, P.; Del Favero, E.; Cantù, L.; Nicoli, S. Polymeric Micelles in Drug Delivery: An Insight of the Techniques for Their Characterization and Assessment in Biorelevant Conditions. J. Control Release 2021, 332, 312–336.
- Lazim, A.M.; Sonthanasamy, R.S.A.; Ling, T.L. Binding Study of Carbon Dots to Bovine Serum Albumin. AIP Conf. Proc. 2019, 2111, 50012.
- Xiao, Q.; Tu, X.; Cao, H.; Luo, H.; Li, B.; Liu, J.; Liu, Y.; Huang, S. Interaction Thermodynamics Investigation of Bovine Serum Albumin with Black Phosphorus Quantum Dots via Spectroscopic and Molecular Simulation Techniques. J. Mol. Struct. 2023, 1276, 134725.
- Preeyanka, N.; Akhuli, A.; Dey, H.; Chakraborty, D.; Rahaman, A.; Sarkar, M. Realization of a Model-Free Pathway for Quantum Dot-Protein Interaction Beyond Classical Protein Corona or Protein Complex. Langmuir 2022, 38, 10704–10715.
- Wang, Z.; Zhao, Q.; Cui, M.; Pang, S.; Wang, J.; Liu, Y.; Xie, L. Probing Temperature- and PH-Dependent Binding between Quantum Dots and Bovine Serum Albumin by Fluorescence Correlation Spectroscopy. Nanomaterials 2017, 7, 93.
- Wang, H.; Shang, L.; Maffre, P.; Hohmann, S.; Kirschhöfer, F.; Brenner-Weiß, G.; Nienhaus, G.U. The Nature of a Hard Protein Corona Forming on Quantum Dots Exposed to Human Blood Serum. Small 2016, 12, 5836–5844.
- Akhuli, A.; Chakraborty, D.; Agrawal, A.K.; Sarkar, M. Probing the Interaction of Bovine Serum Albumin with Copper Nanoclusters: Realization of Binding Pathway Different from Protein Corona. Langmuir 2021, 37, 1823–1837.
- Garon, E.B.; Marcu, L.; Luong, Q.; Tcherniantchouk, O.; Crooks, G.M.; Koeffler, H.P. Quantum Dot Labeling and Tracking of Human Leukemic, Bone Marrow and Cord Blood Cells. Leuk. Res. 2007, 31, 643.
- Wu, X.; Liu, H.; Liu, J.; Haley, K.N.; Treadway, J.A.; Larson, J.P.; Ge, N.; Peale, F.; Bruchez, M.P. Immunofluorescent Labeling of Cancer Marker Her2 and Other Cellular Targets with Semiconductor Quantum Dots. Nat. Biotechnol. 2003, 21, 41–46.
- Nguyen, A.T.; Baucom, D.R.; Wang, Y.; Heyes, C.D. Compact, Fast Blinking Cd-Free Quantum Dots for Super-Resolution Fluorescence Imaging. Chem. Biomed. Imaging 2023, 1, 251–259.
- Hatakeyama, H.; Nakahata, Y.; Yarimizu, H.; Kanzaki, M.; Grünwald, D.; Spottke, B.; Buschmann, V.; Kubitscheck, U.; Lippincott-Schwartz, M.E.J.; Crane, J.M.; et al. Live-cell single-molecule labeling and analysis of myosin motors with quantum dots. Mol. Biol. Cell 2017, 28, 173–181.
- Zhang, M.Q.; Wang, Z.G.; Fu, D.D.; Zhang, J.M.; Liu, H.Y.; Liu, S.L.; Pang, D.W. Quantum Dots Tracking Endocytosis and Transport of Proteins Displayed by Mammalian Cells. Anal. Chem. 2022, 94, 7567–7575.
- Wu, T.; Liu, Y.; Ali, N.M.; Zhang, B.; Cui, X. Effects of Exosomes on Tumor Bioregulation and Diagnosis. ACS Omega 2023, 8, 5157–5168.
- Thakur, A.; Johnson, A.; Jacobs, E.; Zhang, K.; Chen, J.; Wei, Z.; Lian, Q.; Chen, H.J. Energy Sources for Exosome Communication in a Cancer Microenvironment. Cancers 2022, 14, 1698.
- Li, Q. Role of Exosomes in Cellular Communication between Tumor Cells and the Tumor Microenvironment. Oncol. Lett. 2022, 24, 240.
- Jin, Y.; Du, N.; Huang, Y.; Shen, W.; Tan, Y.; Chen, Y.Z.; Dou, W.T.; He, X.P.; Yang, Z.; Xu, N.; et al. Fluorescence Analysis of Circulating Exosomes for Breast Cancer Diagnosis Using a Sensor Array and Deep Learning. ACS Sens. 2022, 7, 1524–1532.
- Zhang, M.; Vojtech, L.; Ye, Z.; Hladik, F.; Nance, E. Quantum Dot Labeling and Visualization of Extracellular Vesicles. ACS Appl. Nano Mater. 2020, 3, 7211–7222.
- Jiang, X.; Zong, S.; Chen, C.; Zhang, Y.; Wang, Z.; Cui, Y. Gold-Carbon Dots for the Intracellular Imaging of Cancer-Derived Exosomes. Nanotechnology 2018, 29, 175701.
- Beltraminelli, T.; Perez, C.R.; De Palma, M. Disentangling the Complexity of Tumor-Derived Extracellular Vesicles. Cell Rep. 2021, 35, 108960.
- Voura, E.B.; Jaiswal, J.K.; Mattoussi, H.; Simon, S.M. Tracking Metastatic Tumor Cell Extravasation with Quantum Dot Nanocrystals and Fluorescence Emission-Scanning Microscopy. Nat. Med. 2004, 10, 993–998.
More
