Spermatogenesis is a very complex process with an intricate transcriptional regulation. The transition from the diploid to the haploid state requires the involvement of specialized genes in meiosis, among other specific functions for the formation of the spermatozoon. The transcription factor cAMP-response element modulator (CREM) is a key modulator that triggers the differentiation of the germ cell into the spermatozoon through the modification of gene expression. CREM has multiple repressor and activator isoforms whose expression is tissue-cell-type specific and tightly regulated by various factors at the transcriptional, post-transcriptional and post-translational level. The activator isoform CREMτ controls the expression of several relevant genes in post-meiotic stages of spermatogenesis. In addition, exposure to xenobiotics negatively affects CREMτ expression, which is linked to male infertility. On the other hand, antioxidants could have a positive effect on CREMτ expression and improve sperm parameters in idiopathically infertile men.
- CREM
- spermatogenesis
- male fertility
- gene regulation
- CREM isoforms
1. Introduction
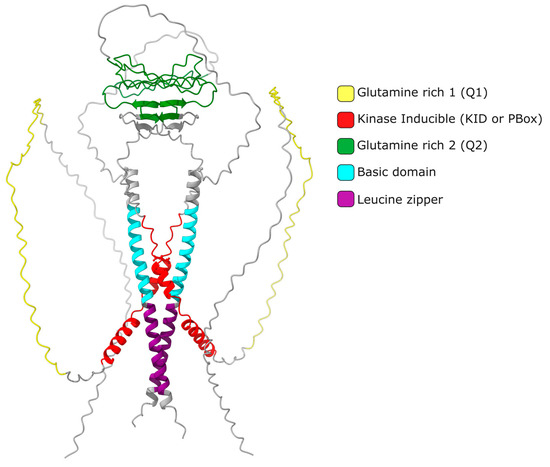
2. CREMτ Expression
It is widely accepted that the up regulation of Crem expression during spermatogenesis occurs in response to LH and FSH signaling[8]. These hormones bind to specific receptors on Leydig and Sertoli cells, leading to an increase in cAMP levels and subsequent changes in gene expression. FSH and forskolin induce an elevated expression of Icer (Inducible cAMP Early Repressor) in rat Sertoli cells. When the ICER protein level increases, it undergoes autoregulation, resulting in a decrease in its expression [17]. Repressor isoforms are initially expressed at low levels in early spermatogonia and spermatocytes. The expression of the activating isoform Cremτ in spermatocytes triggers the expression of meiotic genes crucial for gamete differentiation into spermatozoa. The human CREM gene is located on chromosome 10 (86,108 bp) and consists of eight exons, of which seven exons correspond to the coding sequence of the mRNA (2643 bp) encoding the isoform CREMτ (332 amino acids). In contrast, the mouse Crem gene is located on mouse chromosome 18 (74,504 bp) and contains nine exons, producing a 2362 bp mRNA for the 357aa CREMτ isoform. The mouse Crem gene shares an 86.03% identity with the human gene and encodes a CREMτ protein that shares an 86.63% identity with its human counterpart. To date, 54 transcript isoforms of the human CREM gene, independently of a genome build, have been identified that produce mRNAs ranging from 925 bp to 3500 bp. However, only 17 isoforms and proteins are recognized in specific genomic databases. In contrast, the murine Crem gene has 42 transcript isoforms in the annotated genome GRCm39 C57BL/6J with 18 confirmed isoforms ranging from 529 bp to 2931 pb (Figure 2).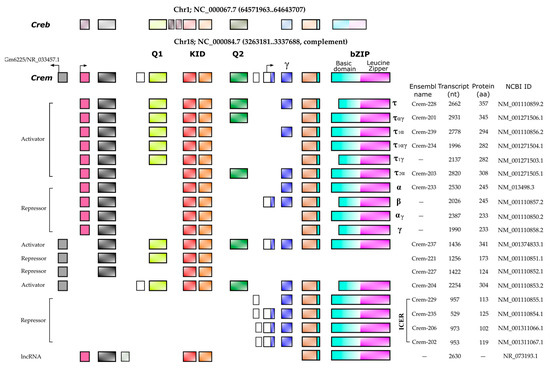
3. CREM Regulation
CREM is widely recognized as a crucial regulator of spermatogenesis; therefore, the comprehension of its modulation at all levels is relevant. A well-established aspect is the direct regulation of CREM activity by ACT-KIFL7b, while the involvement of other ACT-like factors is still largely unclear (see [16] for the latest review). This section focuses on analyzing recent reports on new CREM modulators at transcriptional, post-transcriptional and post-translational levels and their mechanisms, which are summarized in Figure 3.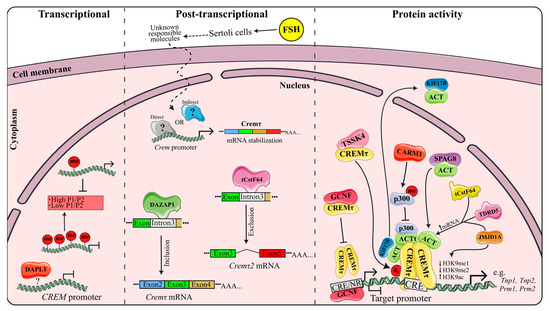
4. Genes Regulated by CREM
5. Male Fertility and CREM
Two research groups in 1996, one led by Sassone-Corsi and the other by Günther Schütz, concurrently described the first mouse model lacking Crem; both studies provided compelling evidence for the impact of a CREM deficiency on spermatogenesis, including the loss of the expression of vital genes for spermatogenesis, arrest at the initiation of spermiogenesis and the absence of sperm production[28][29]. The transcription factor CREM plays a central role in the formation of male haploid cells, as it is responsible for activating genes Involved in meiotic machinery and spermatogenic cell morphogenesis. In a mouse obesity model, a decrease in Crem, adaptor protein 1 (Sh2b1), desert hedgehog (Dhh), insulin-like growth factor 1 (Igf1) and leptin receptor (Lepr) transcript levels was observed, resulting in impaired fertility, such as a reduced sperm motility and decreased mating rates of obese males with female mice [30]. Consequently, it is plausible that CREM homologs may play a similar fundamental role in fertility in other species. Male fertility also relies on the presence of several CREM cofactors, including ACT, KIF17b and SPAG48, which are crucial for the successful development of spermatogenic cells [12][31][32]. Studies in infertile populations have revealed the absence or low expression of CREM or its cofactors, contributing to infertility. In the case of the CREM cofactor ACT, studies in infertile patients with azoospermia or oligozoospermia have identified specific single nucleotide polymorphisms (SNPs) resulting in amino acid changes in the ACT coding region, that, combined to the haplotype 204G-211V-243R-12065G, reduce the interaction between ACT and CREM by 45%, as observed in an in vitro double hybrid test [33]. Similarly, various SNPs found in CREM and its cofactors have been linked to non-obstructive azoospermia (NOA) in patients. SNP rs4934540 in an intron region with a TT or CT genotype confers susceptibility to NOA, while a CT or CC genotype in SNP rs4934540 and an AG genotype of rs2295415 (at the 3′ untranslated end of CREM) reduce the NOA risk. The combination of four CREM SNPs in different haplotypes provides either protection (CGTG) or a high risk (TATG) for spermatogenic failure, as confirmed with expression assays, which showed a low CREM expression[34]. Patients with Klinefelter syndrome commonly experience hypogonadism, low testosterone levels and fertility problems. Testicular biopsies from these patients with mature sperm have demonstrated a lower expression of genes such as CREM and CSF-1, as well as an absence or reduced expression of protamine compared to azoospermic patients without Klinefelter syndrome but in whom complete spermatogenesis was observed histologically [35]. Most Klinefelter patients with SCO show no CREM and protamine expression with CSF-1 expression; however, CREM and protamine levels are still detectable in some patients[35]. Furthermore, alterations in the expression of activating isoforms of CREMτ, namely CREMτ1 and CREMτ2 from the P3 and P4 promoters, respectively, have been observed in patients with spermatogenesis arrest and testicular tumors [36]. Likewise, the involvement of CREM and ACT in the occupation of CRE sites in the promoters of the target genes TNP1 and 2, as well as PRM 1 and 2, in patients with the arrest of round spermatid maturation (SMA) compared to obstructive azoospermia (OA) as a positive control was evaluated. Both CREM and ACT are down-regulated in the group of SMA patients, and low levels of expression in the target genes TNP 1 and 2 and PRM 1 and 2 were also found. A low occupancy of CREM and ACT was also observed in the promoter regions of the TNP1 and 2 and PRM1 and 2 genes in the SMA group, but it could be a result of the low CREM and ACT expression [26]. This confirms CREM’s role as a transcription factor in the development of human spermatogenesis and fertility. Moreover, the CREM promoter contains two CpG islands with a total of 73 CpG sites, making it susceptible to methylation. Differentially methylated sites in CREM have been associated with an impaired sperm DNA integrity in infertile patients [37]. However, bisulfite sequencing studies did not find significant differences in CREM promoter methylation in patients with oligozoospermia or abnormal protamination levels, except for two infertile patients who exhibited a distinct pattern of high methylation[38]. In contrast, a separate pyrosequencing study revealed high levels of DNA methylation in the CREM promoter in patients with oligozoospermia and abnormal protamination, with a negative correlation to the sperm count, morphology and motility[38]. Infection with Toxoplasma gondii has been shown to decrease the reproductive capacity of mice, leading to a noticeable decline in sperm production. An analysis of global methylation in the testicular tissue of infected animals revealed slight differences in methylation levels at specific sites in the Crem promoter [39]. These methylation-prone sites in the CREM promoter can modulate its expression in response to environmental or genotoxic factors, emphasizing their significance as critical determinants of sperm fertility.6. Impact of Xenobiotics in CREMτ Regulation
Previously, the transcriptional regulation of Cremτ was discussed. However, diverse studies suggest that CREMτ is also subject to regulation by xenobiotics. Even though these foreign molecules seem to exert both positive and negative effects on fertility [40][41] extensive research needs to be performed to corroborate if the effect shown in CREMτ levels is due to a lack of a cell population expressing CREM or the direct regulation of the CREM transcript or protein.6.1. CREM Disruption by ROS Production Xenobiotics
In recent years, there has been increasing evidence linking environmental pollutants to semen quality and spermatogenesis impairment, leading to a rise in male infertility rates [42][43]. Several studies have suggested that this effect is due to a down-regulation of CREM pollutants such as solvents, fluoride, pesticides and silica nanoparticles (SiNPs) that seem to be negative regulators of CREM expression[43][44][45]. Common pollutants like 1,2-dichloroethane (1,2-DCE), a widely used solvent, and fluoride, a prevalent contaminant in industrialized countries’ drinking water, seem to reduce CREM expression and its coactivator ACT in a dose-dependent manner. Interestingly, both pollutants are also associated with a decreased semen quality, including increased sperm malformation and a reduced concentration, viability and motility[44][45] Additionally, 1,2-DCE exposures have been linked to the vacuolar degeneration of germ cells, sloughing of spermatogenic cells and increased apoptosis in the testes, resembling the phenotype observed in CREM-knockout mice[28][29]. The pesticide carbendazim (CBZ), another commonly used chemical, has been found to down-regulate CREM expression and decrease sperm concentration and motility. Notably, CBZ disrupts epigenetic markers such as H3K27, 5mC and 5hmC, suggesting that certain pollutants may alter the epigenetic regulation of genes involved in spermatogenesis [45]. Similarly, silica nanoparticles have been shown to reduce sperm quantity and quality by impairing the epigenetic regulation of spermatogenesis and inducing the hypermethylation of the CREM promoter, resulting in CREM down-regulation[38]. It is worth noting that not only pollutants but also certain medical treatments can negatively impact CREM expression. Cistanches herba (CH), a tonic commonly used in Eastern societies, has been found to down-regulate CREM expression in a dose-dependent manner while reducing the testosterone levels, sperm count and sperm motility, mirroring the phenotype observed in pollutant-induced CREM down-regulation [46]. Furthermore, radiation therapy, a known treatment with detrimental effects on the male reproductive system, including permanent infertility, has been shown to decrease CREM expression, decrease testis weight and induce atrophic seminiferous tubules [47]. The down-regulation of CREM by various compounds, whether pollutants or medical treatments, can impair spermatogenesis and lead to male infertility. These xenobiotics likely exert their effects through the production of high levels of reactive oxygen species (ROS), which are associated with testicular damage and male infertility (reviewed in [48]) (Figure 4A). Nevertheless, further research needs to be performed to unravel if the deregulation of CREM by these xenobiotics is because of a down-regulation of Crem per se or due to a lowering of cell populations expressing Crem.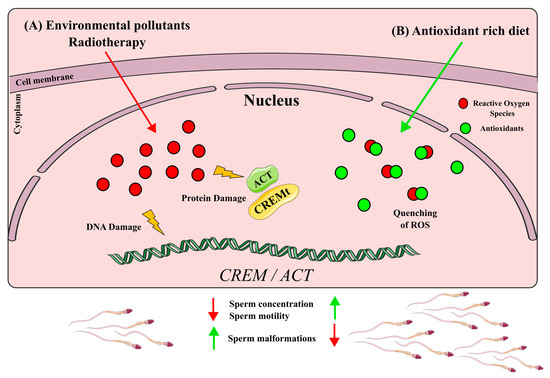
6.2. CREM Up-Regulation by Antioxidants
The up-regulation of CREMτ has been associated with improved semen quality parameters, including sperm concentration and motility [36][30][33]. Traditional Eastern herbal remedies are promising since they have shown a positive impact on CREMτ expression. For example, Rubi Fructus (RF), derived from the dried fruit of Rubus coreanus; Yukmijihwang-tang (YJT), a multiherbal formula used to address male reproductive issues; and MYOMI, a Korean herbal medicine, have traditionally been used to enhance male fertility [49],[50],[51]. Studies administering these formulations to mice have demonstrated an increased sperm concentration and motility, accompanied by an enhancement in Cremτ mRNA and protein levels [51][52]. RF constituents are known for their antioxidant properties[48][52][53][54], while YJT and MYOMI, when tested in combination with cyclophosphamide, a commonly used chemotherapy drug, exhibited a reduced lipid peroxidation, indicating an antioxidant effect [55][56]. Antioxidant supplementation has been found to have a protective effect on male fertility [57] For instance, lutein administration following testicular torsion reduced morphological damage to seminiferous tubules and alleviated testicular oxidative stress. Furthermore, Cremτ expression was restored in lutein-treated mice subjected to testicular torsion[58]. Another essential antioxidant, folic acid, a B-complex vitamin, enhanced semen quality in older roosters. Aging is known to be a factor contributing to male infertility, and folic acid supplementation increased semen volume, sperm concentration and sperm motility in older roosters. Additionally, it led to enhanced mRNA levels of important genes involved in spermatogenesis, including Cremτ [59]. Moreover, antioxidant supplementation in patients with idiopathic infertility resulted in the activation of proteins related to the CREM signaling pathway, such as protein kinase cAMP-dependent regulatory subunits (PRKAR1A, PRKAR2A and PRKACA) and lactate dehydrogenase C (LDHC) [60] Taken together, these studies suggest an improvement in semen parameters and Crem levels due to the ability of antioxidants to quench ROS produced by different types of stress (Figure 4B)7. Other CREM Implications in Health and Disease
As previously mentioned, Cremτ serves as a crucial regulator of spermatogenesis, but it also plays a role in various other molecular mechanisms. Several Crem isoforms have been identified, each with distinct functions beyond spermatogenesis. Repressive isoforms of Crem are involved in the regulation of genes associated with brain function, β cells and immune responses. One such isoform, ICER, participates in numerous neurological processes, including long-term memory, neuronal plasticity, apoptosis and epileptogenesis [61][62][63]. Notably, ICER interacts with brain-derived neurotrophic factor (BDNF) and the signal transducer activator of transcription (STAT3) to bind to pCREB in the Gaba α1 promoter, thereby repressing its transcription in cortical neurons[64][65]. Additionally, ICER modulates adipokine production by inhibiting Creb and suppressing negative effectors of adiponectin, facilitated glucose transporter 4 (GLUT4) and activating transcription factor 3 (ATF3) in adipocytes [66][67]. Another metabolic pathway regulated by ICER is insulin production and secretion. ICER binds to the promoters of genes involved in the insulin pathway, and an increased ICER expression induced by oxidized LDL inhibits insulin production and secretion [68][69]. Furthermore, ICER plays a role in mediating the circadian rhythm in the liver by repressing the period circadian regulator (Per1) gene promoter [70]. It is also implicated in vascular smooth muscle cell apoptosis and proliferation[71]. Another repressive isoform, CREMα, is primarily involved in regulating genes related to the immune system. CREMα acts as a negative regulator of the Cd68 gene promoter and the interleukin Il-2 gene. Surprisingly, it increases the expression of Il17a through epigenetic remodeling [72][73]. CREMα is implicated in immune disorders such as Systemic Lupus Erythematosus (SLE), where its expression is enhanced by transcription factor SP1 and histone lysine methyltransferase SET1 binding to the Crem P1 promoter. This results in increased H3K4me3, decreased DNA methyltransferase 3a (DNMT3a) and the subsequent CREM methylation promoter, leading to the over-expression of CREMα [73][74]. CREMα also binds to the Il17f promoter and represses its expression [75], contributing to accelerated inflammation and autoimmunity [75][76]. In summary, CREM isoforms play diverse roles in various signaling pathways associated with health and disease. However, further extensive research is necessary to fully understand the involvement of other CREM isoforms in different signaling pathways.References
- Kendall, S.K.; Samuelson, L.C.; Saunders, T.L.; et al. Targeted disruption of the pituitary glycoprotein hormone alpha-subunit produces hypogonadal and hypothyroid mice. . Genes Dev. 1995, 9(16), 2007-2019.
- de Kretser, D.M.; Loveland, K.L.; Meinhardt, A.; et al. Spermatogenesis.. Hum. Reprod. 1998, 13(suppl.1), 1-8.
- Griswold, M.D. Cellular and molecular basis for the action of retinoic acid in spermatogenesis. . J. Mol. Endocrinol. 2022, 69(4), T51-T57.
- Schultz, N.; Hamra, F.K.; Garbers, D.L. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. . Proc. Natl. Acad. Sci. USA 2003, 100(21), 12201-12206.
- Shima, J.E.; McLean, D.J.; McCarrey, J.R.; et al. The murine testicular transcriptome: Characterizing gene expression in the testis during the progression of spermatogenesis. . Biol. Reprod. 2004, 71(1), 319-330.
- Foulkes, N.S.; Mellström, B.; Benusiglio, E.; et al. Developmental switch of CREM function during spermatogenesis: From antagonist to activator.. Nature 1992, 355(6355), 80-84.
- Don, J.; Stelzer, G. The expanding family of CREB/CREM transcription factors that are involved with spermatogenesis.. Mol. Cell. Endocrinol. 2002, 187(1-2), 115-124.
- Foulkes, N.S.; Schlotter, F.; Pévet, P.; et al. Pituitary hormone FSH directs the CREM functional switch during spermatogenesis.. Nature 1993, 362(6417), 264-267.
- Foulkes, N.S.; Borrelli, E.; Sassone-Corsi, P. CREM gene: Use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcription. Cell 1991, 64(4), 739-749.
- Behr, R.; Weinbauer, G.F. cAMP response element modulator (CREM): An essential factor for spermatogenesis in primates? . Int. J. Androl 2001, 24(3), 126-135.
- De Cesare, D.; Fimia, G.M.; Sassone-Corsi, P. Signaling routes to CREM and CREB: Plasticity in transcriptional activation. . Trends. Biochem. Sci. 1999, 24(7), 281-285.
- Fimia, G.M.; De Cesare, D.; Sassone-Corsi, P. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature 1999, 398(6723), 165-169.
- Fimia, G.M.; Morlon, A.; Macho, B.; et al. Transcriptional cascades during spermatogenesis: Pivotal role of CREM and ACT. . Mol. Cell. Endocrinol. 2001, 179(1-2), 17-23.
- Macho, B.; Brancorsini, S.; Fimia, G.M.; et al. CREM-dependent transcription in male germ cells controlled by a kinesin. . Science 2002, 298(5602), 2388-2390.
- Kotaja, N.; De Cesare, D.; Macho, B.; et al Abnormal sperm in mice with targeted deletion of the act (activator of cAMP-responsive element modulator in testis) gene. . Proc. Natl. Acad. Sci. USA 2004, 101(29), 10620-10625.
- Hogeveen, K.N.; Sassone-Corsi, P. Regulation of gene expression in post-meiotic male germ cells: CREM-signalling pathways and male fertility. . Hum. Fertil. 2006, 9(2), 73-79.
- Monaco, L.; Foulkes, N.S.; Sassone-Corsi, P. Pituitary follicle-stimulating hormone (FSH) induces CREM gene expression in Sertoli cells: Involvement in long-term desensitization of the FSH receptor. . Proc. Natl. Acad. Sci. USA 1995, 92(23), 10673-10677.
- Delmas, V.; Sassone-Corsi, P. The key role of CREM in the cAMP signaling pathway in the testis. . Mol. Cell. Endocrinol. 1994, 100(1-2), 121-124.
- Ruppert, S.; Cole, T.J.; Boshart, M.; Multiple mRNA isoforms of the transcription activator protein CREB: Generation by alternative splicing and specific expression in primary spermatocytes. . EMBO J. 1992, 11(4), 1503-1512.
- Walker, W.H.; Sanborn, B.M.; Habener, J.F. An isoform of transcription factor CREM expressed during spermatogenesis lacks the phosphorylation domain and represses cAMP-induced transcription. . Proc. Natl. Acad. Sci. USA 1994, 91(26), 12423-12427.
- Gene ID 1390 . Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; 2004. Retrieved 2023-10-13
- GENE ID 12916 . Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information; 2004. Retrieved 2023-10-13
- Peri, A.; Krausz, C.; Cioppi, F.; et al Cyclic adenosine 3’,5’-monophosphate-responsive element modulator gene expression in germ cells of normo- and oligoazoospermic men. J. Clin. Endocrinol. Metab 1998, 83(10), 3722-3726.
- Noda, T.; Shidara, O.; Harayama, H.; et al Detection of the activator cAMP responsive element modulator (CREM) isoform ortholog proteins in porcine spermatids and sperm. Theriogenology 2012, 77(7), 1360-1368.
- Jazireian, P.; Favaedi, R.; Sadighi Gilani, M.A.; et al Dynamic Expression and Chromatin Incorporation of ACT and CREM Transcription Factors in Testis Tissues of Infertile Men. . Cell J 2021, 23(7), 736-741.
- Kaprio, H.; Heuser, V.D.; Orte, K.; et al Expression of Transcription Factor CREM in Human Tissues. J. Histochem. Cytochem 2021, 69(8), 495-509.
- Kumar, T. R; Low, M. J.; Matzuk, M. M. Genetic rescue of follicle-stimulating hormone beta-deficient mice. Endocrinology 1998, 139(7), 3289-3295.
- Nantel, F.; Monaco, L.; Foulkes, N.S.; et al Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature 1996, 380(6570), 159-162.
- Martianov, I.; Choukrallah, M.A.; Krebs, A.; et al Cell-specific occupancy of an extended repertoire of CREM and CREB binding loci in male germ cells. BMC Genom 2010, 11, 530.
- Christensen, G.L.; Wooding, S.P.; Ivanov, I.P.; et al Sequencing and haplotype analysis of the activator of CREM in the testis (ACT) gene in populations of fertile and infertile males. Mol. Hum. Reprod 2006, 12(4), 257-262.
- Kotaja, N.; Macho, B.; Sassone-Corsi, P. Microtubule-independent and protein kinase A-mediated function of kinesin KIF17b controls the intracellular transport of activator of CREM in testis (ACT). . J. Biol. Chem 2005, 280(36), 31739-31745.
- Wu, H.; Chen, Y.; Miao, S.; Zhang, C.; et al Sperm associated antigen 8 (SPAG8), a novel regulator of activator of CREM in testis during spermatogenesis. . FEBS Lett 2010, 584(13), 2807-2815.
- He, X.J.; Song, B.; Du, W.D.; et al CREM variants rs4934540 and rs2295415 conferred susceptibility to nonobstructive azoospermia risk in the Chinese population. Biol. Reprod 2014, 91(2), 52.
- Abofoul-Azab, M.; Lunenfeld, E.; Kleiman, S.; et al Determining the expression levels of CSF-1 and OCT4, CREM-1, and protamine in testicular biopsies of adult Klinefelter patients: Their possible correlation with spermatogenesis. Andrologia 2022, 54(10), e14558.
- Song, B.; Wang, C.; Chen, Y.; et al Sperm DNA integrity status is associated with DNA methylation signatures of imprinted genes and non-imprinted genes. J. Assist. Reprod. Genet 2021, 38(8), 2041-2048.
- Grozdanov, P.N.; Amatullah, A.; Graber, J.H.; et al TauCstF-64 Mediates Correct mRNA Polyadenylation and Splicing of Activator and Repressor Isoforms of the Cyclic AMP-Responsive Element Modulator (CREM) in Mouse Testis. Biol. Reprod 2016, 94(2), 34.
- Dvorakova-Hortova, K.; Sidlova, A.; Ded, L.; et al Toxoplasma gondii decreases the reproductive fitness in mice. PLoS ONE 2014, 9(6), e96770.
- Sang, Y.; Liu, J.; Li, X.; et al The effect of SiNPs on DNA methylation of genome in mouse spermatocytes. Environ. Sci. Pollut. Res. Int 2021, 28(32), 43684-43697.
- Boekelheide, K. Mechanisms of toxic damage to spermatogenesis. J. Natl. Cancer. Inst. Monogr 2005, 34, 6-8.
- Chen, H.Y.; Yu, Y.H.; Yen, P.H. DAZAP1 regulates the splicing of Crem, Crisp2 and Pot1a transcripts. Nucleic Acids Res 2013, 41(21), 9858-9869.
- Levine, H.; Jørgensen, N.; Martino-Andrade, A.; et al Temporal trends in sperm count: A systematic review and meta-regression analysis. Hum. Reprod. Update 2017, 23(6), 646-659.
- Centola, G.M.; Blanchard, A.; Demick, J.; et al Decline in sperm count and motility in young adult men from 2003 to 2013: Observations from a U.S. Sperm Bank. Andrology 2017, 4(2), 270-276.
- Zhang, Y.; Li, G.; Zhong, Y.; et al Dichloroethane Induces Reproductive Toxicity Mediated by the CREM/CREB Signaling Pathway in Male NIH Swiss Mice. Toxicol. Sci. 2017, 160(29), 299-314.
- Wang, C.; Chen, Y.; Manthar, R.K.; et al Abnormal spermatogenesis following sodium fluoride exposure is associated with the downregulation of CREM and ACT in the mouse testis. Toxicol. Sci 2017, 160(2), 299-314.
- Liu, J.; Zhang, P.; Zhao, Y.; et al Low dose carbendazim disrupts mouse spermatogenesis might Be through estrogen receptor related histone and DNA methylation. Ecotoxicol. Environ. Saf 2019, 176, 242-249.
- Kim, S.W.; Yoo, S.H.; Lee, H.J.; et al Cistanches herba induces testis cytotoxicity in male mice. Bull. Environ. Contam. Toxicol 2012, 88(1), 112-117.
- Nagahori, K.; Qu, N.; Kuramasu, M.; et al Changes in Expression of Specific mRNA Transcripts after Single- or Re-Irradiation in Mouse Testes. Genes 2022, 13(19), 151.
- Guerriero, G.; Trocchia, S.; Abdel-Gawad, F.K.; Ciarcia, G. Roles of reactive oxygen species in the spermatogenesis regulation.. Front. Endocrinol 2014, 5, 56.
- Kim, E.J.; Lee, Y.J.; Shin, H.K.; Park, J.H. Induction of apoptosis by the aqueous extract of Rubus coreanum in HT-29 human colon cancer cells.. Nutrition 2005, 21(11-12), 1141-1148.
- Bensky, D.; Barolet, R. . Chinese Herbal Medicine Formulas and Strategies; Eastland Press: Seattle, WA, USA, 1990; pp. 263-264.
- Pang, G.C.; Kim, M.S.; Lee, M.W. Hydrolyzable tannins from the fruits of Rubus coreanum. Korean J. Pharmacogn 1996, 27(4), 366-370.
- Kim, S.W.; Shin, M.H.; Jung, J.H. et al A triterpene glucosyl ester from the roots of Rubus crataegifolius. . Arch. Pharmacal Res 2001, 24(5), 412-415.
- Patel, A.V.; Rojas-Vera, J.; Dacke, C.G. Therapeutic Constituents and Actions of Rubus Species. Curr. Med. Chem. 2004, 11(11), 1501-1512.
- Oh, M.S.; Yang, W.M.; Chang, M.S. et al Effects of Rubus coreanus on sperm parameters and cAMP-responsive element modulator (CREM) expression in rat testes. J. Ethnopharmacol 2007, 114(3), 463-467.
- Oh, M.S.; Chang, M.S.; Park, W. et al Yukmijihwang-tang protects against cyclophosphamide-induced reproductive toxicity. Reprod. Toxicol 2007, 24(3-4), 365-370.
- Arafa, M.; Agarwal, A.; Majzoub, A. et al Efficacy of antioxidant supplementation on conventional and advanced sperm function tests in patients with idiopathic male infertility.. Fertil. Steril. 2019, 112(39), e362.
- Al-Maghrebi, M.; Renno, W.M.; Al-Somali, H.F. et al Lutein modulates transcription dysregulation of adhesion molecules and spermatogenesis transcription factors induced by testicular ischemia reperfusion injury: It could be SAFE. Naunyn-Schmiedeberg’s Arch. Pharmacol 2016, 389(5), 539-551.
- Ye, N.; Lv, Z.; Huang, Z. et al Dietary folic acid supplementation improves semen quality and spermatogenesis through altering autophagy and histone methylation in the testis of aged broiler breeder roosters.. Theriogenology 2022, 181, 8-15.
- Agarwal, A.; Panner Selvam, M.K.; Samanta, L. et al Effect of Antioxidant Supplementation on the Sperm Proteome of Idiopathic Infertile Men.. Antioxidants 2019, 8(10), 488.
- Mioduszewska, B.; Jaworski, J.; Szklarczyk, A.W. et al Inducible cAMP early repressor (ICER)-evoked delayed neuronal death in the organotypic hippocampal culture. J. Neurosci. Res 2008, 86(1), 61-70.
- Kojima, N.; Borlikova, G.; Sakamoto, T. et al . Inducible cAMP early repressor acts as a negative regulator for kindling epileptogenesis and long-term fear memory. J. Neurosci 2008, 28(25), 6459-6472.
- Porter, B.E.; Lund, I.V.; Varodayan, F.P. et al The role of transcription factors cyclic-AMP responsive element modulator (CREM) and inducible cyclic-AMP early repressor (ICER) in epileptogenesis. Neuroscience 2008, 152(3), 829-836.
- Hu, Y.; Lund, I.V.; Gravielle, M.C. et al Surface expression of GABAA receptors is transcriptionally controlled by the interplay of cAMP-response element-binding protein and its binding partner inducible cAMP early repressor. J. Biol. Chem 2008, 283(14), 9328-9340.
- Lund, I.V.; Hu, Y.; Raol, Y.H. et al BDNF selectively regulates GABAA receptor transcription by activation of the JAK/STAT pathway.. Sci. Signal 2008, 1(41), ra9.
- Favre, D.; Le Gouill, E.; Fahmi, D. et al Impaired expression of the inducible cAMP early repressor accounts for sustained adipose CREB activity in obesity. Diabetes 2011, 60(12), 3169-3174.
- Brajkovic, S.; Marenzoni, R.; Favre, D. et al Evidence for tuning adipocytes ICER levels for obesity care. . Adipocyte 2012, 1(3), 157-160.
- Abderrahmani, A.; Cheviet, S.; Ferdaoussi, M. et al ICER induced by hyperglycemia represses the expression of genes essential for insulin exocytosis.. EMBO J 2006, 25(5), 977-986.
- Favre, D.; Niederhauser, G.; Fahmi, D. et al Role for inducible cAMP early repressor in promoting pancreatic beta cell dysfunction evoked by oxidative stress in human and rat islets. Diabetologia 2011, 54(9), 2337-2346.
- Zmrzljak, U.P.; Korenčič, A.; Košir, R. et al Inducible cAMP early repressor regulates the Period 1 gene of the hepatic and adrenal clocks.. J. Biol. Chem 2013, 288(15), 10318–10327.
- Ohtsubo, H.; Ichiki, T.; Miyazaki, R. et al Inducible cAMP early repressor inhibits growth of vascular smooth muscle cell. Arter. Thromb. Vasc. Biol 2007, 27(7), 1549-1555.
- Ahlmann, M.; Varga, G.; Sturm, K. et al The cyclic AMP response element modulator {alpha} suppresses CD86 expression and APC function. J. Immunol. 2009, 182(7), 4167-4174.
- Hedrich, C.M.; Crispin, J.C.; Rauen, T. et al cAMP response element modulator α controls IL2 and IL17A expression during CD4 lineage commitment and subset distribution in lupus. . Proc. Natl. Acad. Sci. USA 2012, 109(41), 16606-16611.
- Juang, Y.T.; Rauen, T.; Wang, Y.; Ichinose, K.; Benedyk, K.; Tenbrock, K.; Tsokos, G.C. Transcriptional activation of the cAMP-responsive modulator promoter in human T cells is regulated by protein phosphatase 2A-mediated dephosphorylation of SP-1 and reflects disease activity in patients with systemic lupus erythematosus. J. Biol. Chem. 2011, 286, 1795–1801.
- Zhang, Q.; Ding, S.; Zhang, H.; Long, H.; Wu, H.; Zhao, M.; Chan, V.; Lau, C.S.; Lu, Q. Increased Set1 binding at the promoter induces aberrant epigenetic alterations and up-regulates cyclic adenosine 5′-monophosphate response element modulator alpha in systemic lupus erythematosus. Clin. Epigenet. 2016, 8, 126.
- Hedrich, C.M.; Rauenl, T.; Kis-Tothl, K.; Kyttaris, V.C.; Tsokos, G.C. cAMP-responsive element modulator α (CREMα) suppresses IL-17F protein expression in T lymphocytes from patients with systemic lupus erythematosus (SLE). J. Biol. Chem. 2012, 287, 4715–4725.
- Lippe, R.; Ohl, K.; Varga, G.; Rauen, T.; Crispin, J.C.; Juang, Y.T.; Kuerten, S.; Tacke, F.; Wolf, M.; Roebrock, K.; et al. CREMα overexpression decreases IL-2 production, induces a T(H)17 phenotype and accelerates autoimmunity. J. Mol. Cell. Biol. 2012, 4, 121–123.
