You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 1 by Ramesh Namdeo Pudake and Version 2 by Jason Zhu.
It is crucial to increase agricultural yields to fulfill the rising demand for food and the security it provides for a growing population. To protect human food supplies and agricultural outputs, disease management is essential. Plant infections are a silent enemy of economic crop production and cross-border commerce of agricultural goods, inflicting roughly 20–30% losses a year. If infections are accurately and rapidly detected and identified, this can be minimized, and specialized treatment can be given.
- nanotechnology
- sustainability
- plant disease
- detection
1. Introduction
The primary reasons for limiting agricultural productivity are pathogen infections, which have become one of the key problems in the global scenario [1][47]. Despite being excessively slow and unsuited for general use, traditional methods for detecting pathogens and diagnosing plant diseases are typically only somewhat accurate. To identify plant pathogenic organisms with a high degree of accuracy and precision, traditional molecular diagnostic techniques, including polymerase chain reaction (PCR), enzyme-linked immunosorbent assay (ELISA), and other established techniques such as colony counting, fluorescence in situ hybridization (FISH), etc., are frequently used in laboratories around the world [2][48]. However, the use of these conventional methods in underdeveloped nations is constrained due to the demand for specialized equipment, laboratory setup, and experts to handle the equipment [3][49]. The application of nanotechnology in plant disease diagnostics might revolutionize research and lead to the development of cutting-edge instruments for the early and quick detection of plant infections. Nanomaterials are excellent options for this purpose because of their size (1–100 nm), which can offer improved surface-to-volume ratios and have exclusive chemical, photosensitive, and electrical properties that are not present in their bulk equivalents [4][50]. The ability to modify molecules at the nanoscale and the unique optical properties provided by nanomaterials will enable highly sensitive and useful detection of plant pathogens [5][6][4,51]. Biosensors, with the help of nanoparticles, can show better performance in selectivity, sensitivity, and detection limits. It is also possible to miniaturize the devices of various nanoparticles because of their small size. The nanoparticles provide advantages such as increased conductivity in the sensing platform; a high surface-to-volume ratio that increases the binding/immobilization surface for the bioreceptors; and tuning of the surface moieties on nanoparticles to create binding sites for biomolecule immobilization. Along with this, the use of molecularly imprinted polymers (MIPs) and DNA-based aptamers makes the biosensors more stable in all conditions. These properties make nanosensors a better alternative to conventional techniques [7][52]. Various applications of nanotechnology in plant disease detection are discussed herewith (Table 1, Figure 1).
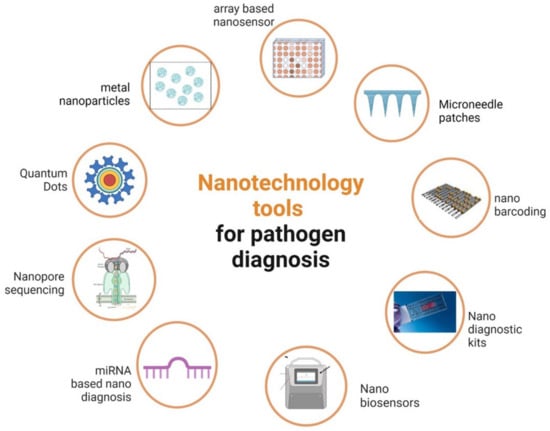
Figure 1.
Diagrammatic representation of different nanotechnology tools for phytopathogen detection.
Table 1.
Nanotechnology-based diagnosis of plant pathogens/diseases.
| Nanomaterial/ Substrate |
Disease/Causal Organism | Target Plants | Bio-Recognition | Detection, LOD/Accuracy | References |
|---|---|---|---|---|---|
| SWCNTs/Gold microelectrodes on a Si/SiO2 wafer |
Sec-delivered effector 1 (SDE1) |
Citrus | Anti-SDE1 polyclonal | FET/LOD: 5 nM | [8][53] |
| Au and pentacene films/Gold gate electrode | PPV | Stone fruit trees |
Anti-Plum Pox Virus polyclonal |
EGOFET/LOD: 180 pg mL−1 |
[9][54] |
| SWCNTs/Silicon wafer covered with SiO2 | p-Ethylphenol released by Phytophthora |
Strawberry | ssDNA | E-nose 0.13% of Pethylphenol | [10][55] |
| N- and B-doped MWCNTs/Interdigitated Electroless nickel immersion gold electrodes |
VOCs exhaled by Aspergillus and Rhizopus fungi | Strawberry | - | E-nose | [11][56] |
| rGO and Au NPs/ Kirigami-based structure with AgNW electrodes |
VOCs exhaled by Phytophthora infestans infection | Tomato | Chemiresistive sensor array/ >97% accuracy |
[12][57] | |
| Au NPs/GCE | Xanthomonas axonopodis |
Citrus | Anti-PthA | FET-SWV/LOD: 0.01 nM |
[13][58] |
| Au NPs/SPCE | CTV | Citrus | Thiolated ssDNA |
EIS/LOD: 100 nM | [14][59] |
| Au NPs/SPCE | CTV detection | - | Thiolated primer |
EIS L/OD: 1 pg μL−1 | [15][60] |
| TiO2 and SnO2 nanoparticles/SPCE |
p-ethylguaiacol, volatile compound due to Phytophthora cactorum fungus infection |
CV and DPV/LOD: 35–62 nmol L−1 |
[16][61] | ||
| Au NPs/GCE | PSS | Stewartia | HRP | LSV/7.8 × 103 cfu mL−1 | [17][62] |
| GO/Paper electrodes | False smut caused by Ustilaginoidea virens | Rice | ssDNA | CV and LSV/10 fmol L−1 | [18][63] |
| GO/ITO | GBNV | Groundnut | anti-GBNV | DPV/LOD: 5.7 ng mL−1 |
[19][64] |
| Au NPs/SPCE | Detection of plant pathogen DNA | - | Recombinase polymerase amplification |
DPV/214 pmol L−1 | [20][65] |
| PPY nanoribbon/Gold microelectrode | Cucumber mosaic virus (CMV) | Cucumber | anti-CMV IgG | Chemiresistive Microelectrode/LOD 10 ng ml−1 |
[21][66] |
| Au NPs/SPCE | CMV | Cucumber | - | Chronoamperometry | [22][67] |
| Au NPs/SPCE | Rice tungro disease | Rice | anti- RTBV/ RTSV |
Cyclic voltammtry | [23][68] |
| Au NPs/Carbon ink 8-WE SPCE | CTV | Sweet orange trees |
anti-bodies Ab1 and Ab2 |
Amperometry/LOD: 0.3 fg mL−1 |
[24][69] |
| Au NPs | P. infestans | Tomato | (Cys)-capped | C/0.4 ppm | [25][70] |
| Au NPs | Xanthomonas campestris |
Brassica | Colorimetric/102 CFU mL−1 | [26][71] | |
| Fluorescent nanoparticles |
Phakopsora Pachyrhizi | Soybean | IgG antibodies | fluorescence 2.2 ng mL−1 | [27][72] |
| Au NPs reverse primer (20-mer) | Yellow leaf curl virus | Tomato | Reverse primer (20- mer) | LSPR/5 ng μL−1 |
[28][73] |
| Au NPs-SA | Alternaria panax Whetz | Ginseng | Mouse anti-Fam antibody and BSA-Biotin | LFA/0.01 pg μL−1 |
[29][74] |
| Au NPs | Phytophthora infestan | Potato | Streptavidinbiotinylated T and C |
LFA/0.01 pg μL−1 |
[30][75] |
| Au NPs and silver | Leafroll virus | Potato | Anti-PLRV antibodies | LFA 0.2 ng ml−1 |
[31][76] |
| Carbon nanoparticles |
X. arboricola pv. Pruni | Stone fruits and almond | Polyclonal antibodies 2626.1-WC |
LFA 104 CFU mL−1 |
[32][77] |
2. Bio-Nanomaterials
Bio-nanomaterials have greatly benefited the disciplines of biology, medical sciences, and agriculture. By creating more analytical tools in nanotechnology for accurate environmental hazard management, the use of bio-nanomaterials will be more effective in the environment [33][78]. In addition, pure cultures of microbes or their associated proteins and enzymes, as well as plant extracts, can be employed to bio-synthesize nanomaterials [34][79].
It has been demonstrated that nanotechnology is a key tool for identifying and quantifying plant pathogens. Examples of nanostructures with device miniaturization potential include those utilized to develop biosensors with increased selectivity, sensitivity, and limit of detection for onsite detection [10][55]. Sensor conductivity can be enhanced, for instance, by using carbon nanotubes or graphene derivatives [25][35][70,80]. Surface-functionalized electrospun nanofibers or metallic nanoparticles [36][81] significantly enhance the surface-to-volume ratio, allowing for bio-specific immobilization [13][25][37][58,70,82]. A molecularly imprinted polymer (MIP), which solely interacts with the target analyte and eliminates interferences, can be used to alter the sensor’s selectivity [38][83].
Thermographic and hyperspectral imaging are some of the imaging techniques that have been used in the field for the indirect detection of plant disease. The sensitivity to changes in environmental parameters and the lack of specificity for different strains or disease subtypes are only two of its major shortcomings. Chemo- or biosensing technology has developed quickly, leading to a broad variety of useful applications, such as the assessment of the quality of pharmaceutical and industrial products. The analytes may be recognized by the nanosensors characteristic of electrical or optical outputs, which have recently been proposed and sold for agricultural diagnostics, depending on the transduction processes of the defined sensory interactions. The sensor’s detection specificity may be improved by using selective chemical interactions or biospecies identification tools such as antibodies, enzymes, DNA oligos, and aptamers. To boost the detection sensitivity, surface-enhanced optical characteristics, such as electron-conductive nanoscale substrates or surface plasmon resonance (SPR), including graphene or carbon nanotubes, may be utilized as transducers [39][84].
3. Nanoparticle Bio-Barcode Assay
The bio-barcode test based on nanoparticles is more capable of identifying infections than standard techniques such as ELISA, real-time PCR, etc. [40][85]. It can also aid in the rapid recognition of plant diseases. In the bio-barcode method, magnetic microbeads (MMB) and gold nanoparticles (Au NP) are employed as probes. Many bio-barcode components have also undergone changes to broaden their use. DNA barcoding has been suggested as a method for detecting fungi DNA with a bio-barcode (b-DNA) [41][86]. For amplifying and locating proteins or nucleic acids, it is an exceptionally sensitive approach. In DNA bio-barcoded assays, the target protein is quickly separated from the sample using magnetic gold nanoparticles (Au-MNPs) that have been tailored with oligonucleotides for signal amplification. Considerable signal amplification is made possible by the high b-DNA-to-recognition agent ratio. It is particularly promising because, under ideal circumstances, it allows for the quick detection of a variety of nucleic acids at high zeptomolar levels and protein targets at low attomolar concentrations. A revolutionary approach that could replace the PCR technique is the bio-barcode test [4][50].
4. Nanopore DNA Sequencing
For disease management methods, accurate plant pathogen detection and identification are crucial. Common diagnostic techniques for identifying plant infections have drawbacks, including the need for previous knowledge of the genome sequence, limited sensitivity, and a constrained capacity to identify many diseases at once. The total nucleic acid concentration in biological samples may now be determined thanks to the advancement of DNA sequencing technology. Nanopore devices may be used to examine genetic data quickly and affordably without the requirement for sample preparation. Due to the protein nanopores in their membranes, this device distinguishes between nucleotides by measuring variations in conductivity. A bilayer membrane composed of polymers and nanopores in the chip is tied to a sensor. With the invention of this new technique, it is now feasible to recognize epidemics and track their progress, as well as differentiate between various pathogens, challenge genetic components, and sequence two different genes that are found on the same chromosome. By utilizing a nanopore model that is previously present in a contemporary diagnostic gadget, a whole genome analysis might be completed swiftly. To improve agricultural crops, it may be used to research the genomes of plants and diseases.
Plant pathogens pose a hazard to crop quality and productivity; as a result, effective and precise pathogen diagnostics are essential for managing and controlling crop disease. Research into plant viruses has been transformed by recent developments in sequencing technology. Because of its high sensitivity, high throughput, and lack of sequence dependency, next-generation sequencing (NGS), which represents metagenomics sequencing technology, has significantly advanced the development of viral diagnostics research. However, the expensive cost, labor-intensive nature, and cumbersome equipment of NGS-based viral identification techniques place a limit on their effectiveness. Long DNA or RNA readings may now be directly sequenced in real time, thanks to developments in nanopore sequencing. This is widely utilized in plant virus surveillance, virus discovery, viral genome assembly, and evolutionary research because of its versatility, portable sequencers, and adaptable data analysis [42][87]. Nanopore single-molecule sequencing technology is also being used to diagnose plant bacterial and fungal diseases. It was examined using DNA or RNA that had been obtained from the tissues of plants that had been injected with diseases that cause pathogens and exhibited the symptoms. Using nanopore sequencing, pathogens can be detected in real time and categorized to the species or genus level.
Conventional diagnostic techniques (including PCR, ELISA, and the Koch test) were used to validate DNA sequencing or direct RNA sequencing of samples containing unidentified disease pathogens, which corroborated the outcomes of nanopore sequencing. Long read lengths, quick run times, portability, cheap cost, and the potential for usage in any laboratory are all benefits of this technology [43][88]. The Oxford Nanopore Technologies tool “MinION” is a handheld sequencing system, and it was found to be an efficient method for the diagnosis of various plant pathogens, including fungi such as S. lycopersicum in tomato and P. digitatum in lemon [43][88]. Two pathogens, Candidatus Liberibacter asiaticus and Plum pox virus, were quickly detected (within 24 h) in the peach by [44][89] using nanopore sequencing in conjunction with whole transcriptome amplification. By anticipating the existence of numerous plant viral species, such as Dioscorea bacilliform virus, Yam mild mosaic virus, and Yam chlorotic necrosis virus, in a water yam plant, [45][90] revealed high genome mapping findings attained by MinION. The entire experiment takes less than two hours, and the outcomes are equivalent to those of other diagnostic techniques (such as PCR and ELISA). Even though the present technique still has several drawbacks, such as a high per-read error rate and a limited ability to tell apart similar sequences, additional developments in nanopore technology will lead to the creation of more powerful sequencing platforms [39][84].
5. Nanodiagnostic Kit
A nanodiagnostic kit, often known as a “lab inside a box”, is a small box that is used to monitor important plant processes that may be performed in a small space [46][91]. Several hurdles must be cleared before nanodiagnostic kit-based equipment systems may be used reliably in agriculture and related fields. The diagnostic kits’ specificity may be improved, and strain differentiation can be achieved by several means, one of which is the identification and selection of efficient antigen, antibody, and nucleotide targets. It is also vital to create international standards for assessing tests and detection levels to compare studies on detection limits. Additionally, strategies for streamlining purification and isolating important genes are necessary for identifying the genetic targets of a particular illness [47][92].
These point-of-care kits and devices can assist farmers in limiting the spread of infectious illnesses by quickly identifying plant pathogens [48][49][93,94]. One strip with four mycosensors reportedly has the ability to detect ZEA, T-2/HT-2, DON, and FB1/FB2 mycotoxins in cash crops including wheat, barley, and maize [50][95]. Maize Chlorotic Mottle Virus (MCMV), the only member of the Mahromovirus genus, often co-infects plants with one or more viruses from the Potyvirus genus and presents a significant threat to the global maize economy. The application of viral integrated management techniques requires the swift and precise identification of the disease’s causal agent. Six super-sensitive and precise monoclonal antibodies (mAbs) against MCMV were first developed in one study using pure MCMV virions as the immunogen. Following the discovery of the mAbs, the quantum dot enzyme-linked immunosorbent test (Dot-ELISA) was created, which was capable of detecting MCMV in the crude extract of infected maize leaf. A rapid and easy gold nanoparticle-based immunochromatographic test strip (Au NP-ICTS) based on the paired mAbs 7B12 and 17C4 was further developed to monitor MCMV in point-of-care testing. This test strip can identify the virus in crude extracts of MCMV-infected maize leaves that have been diluted 25,600 times (w/v, g/mL). It took 10 min to complete the whole ICTS test process. When compared to conventional reverse transcription-polymerase chain reaction (RT-PCR), the detection endpoint of both serological methods is higher than that of RT-PCR, notably the Dot-ELISA, which is 12.1 times more sensitive. Additionally, there was concordance between the RT-PCR outcomes and the detection outcomes of 20 blinded maize samples from the two serological assays. The newly created Dot-ELISA and Au NP-ICTS offer tremendous application potential for the detection of MCMV in plant samples [51][96]. Although nanotechnology has not yet been completely utilized to identify pathogen infections in agricultural crops, it has the ability to resolve many of the issues previously mentioned for efficient on-site real-time diagnosis of crop diseases [47][92].
6. Quantum Dots (QDs)
Semiconducting nanocrystals called quantum dots emit certain light wavelengths and change the exposed light spectrum into a distinct frequency of energy. They are three-dimensional nanoparticles with a broad excitation spectrum [52][97]. QD-based nanosensors can be helpful for detecting a number of enzymes and infections [53][54][98,99]. Quantum dots (QDs) are made of elements from the periodic groups II–VI, III–V, or IV–VI that have special photophysical characteristics. They are also referred to as zero-dimensional materials since they are nanostructured materials. One of the most common QDs are cd-chalcogenide nanocrystals, which have a ZnS shell around a centrosome that is 2–10 nm in size. They typically range in size from 10–15 nm when the outside of the shell is covered with a polymer. According to reports, CdS, CdTe, and CdSe are typically employed as the centrosomes of quantum dots [55][100]. Other materials lack the unique photophysical properties of QDs. In contrast to conventional fluorescent probes such as fluorescent proteins and organic dyes, QDs are distinguished by their size-tunable light emissions, limited and symmetric emission spectra, and wide absorption spectra that provide simultaneous stimulation of various fluorescent hues. In addition, compared to other materials, QDs exhibit a remarkable increase in brightness and resistance to photobleaching [56][57][58][101,102,103]. Quantum dot-based biosensors make use of QDs as the interface component and have names such as QD-based BRET immunosensor, QD-based FRET immunosensor, and QD-based FRET genosensor, depending on the kind of molecular beacon attached to the QDs and transduction signals [59][60][104,105]. The conceptual basis of the QD-based FRET genosensor is commonly used in biological applications [54][99].
In one study, glutathione-S-transferase (GST) proteins, which are specific to Polymyxabetae, were detected using CdTe quantum dots coated with antibiotics as biosensors [61][106]. Fluorescence resonance energy transfer (FRET) depends on resonance dipole–dipole coupling, which is produced by rhodamine’s interactions with CdTe quantum dots. In less than 30 min, this device may be used to evaluate plants and produce useful results. In a different investigation, P. aurantifolia was sensitively detected using a QD-based sensor, and the sensor’s 100% specificity was demonstrated in sick lime plants [62][11]. Despite the fact that QD-based biosensors are a relatively new form of sensor and are predicted to provide new possibilities for managing plant diseases, some investigations have also been carried out on other agricultural pathogens by applying QD-based biosensors [63][64][65][107,108,109] for the detection of plant infections. It is plausible to assume that quantum dots will contribute to the impending revolution in plant pathogen detection if their unique photophysical properties are taken into consideration as an interface component [4][50].
