You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Peter Tang and Version 1 by Alice Vilela.
Flavors and fragrances are essential for the beverage and food industries. Biosynthesis or extraction are the two main ways to obtain these crucial compounds with many different chemical structures.
- Natural flavours
- de novo synthesis
- ligand-receptor interaction
- e-nose
- e-tongue
- sensory analysis
1. Introduction
The increased consumer preference for natural and sustainable products makes the production of natural flavors, which define the sensory perception of beverages and other food products, an ever-challenging purpose for academic and industrial investigation [1]. Indeed, olfactory signaling initiates when odorant molecules contact the olfactory sensory neurons, which express proteins that convert chemical signals into neuronal impulses. Consequently, neurochemical effects occur in several brain regions, generating the smell sense. The discovery that each olfactory neuron expressed one particular odorant receptor allowed the researchers Linda Buck and Richard Axel to be distinguished with the Nobel Prize in Medicine in 2004.
Odorant or olfactory receptors (ORs) are located in the human nose, specifically in the olfactory epithelium. However, several studies in recent years have shown the existence of the “ectopic olfactory receptors” in mammals, i.e., receptors of the same type spread in many diverse organs and systems [2] such as in tissues as the testes [3] and heart [4]. ORs and related signaling molecules can also be present in non-olfactory areas of the nervous system and may trigger vulnerability to neurodegenerative diseases and drugs [2].
The olfactory receptors recognize many different odor molecules of a diverse protein sequence. According to Farbiszewski and Kranc [5], the olfactory receptors include 172 subfamilies encoded by a single chromosomal locus. OR genes constitute the most abundant family of G protein-coupled receptors (GPCRs), with about 1000 genes in the mouse [6]. The human receptor gene family comprises 339 receptor genes and 297 receptor pseudogenes, unequally dispersed in 51 distinct loci on 21 human chromosomes [5]. However, almost half of the human OR repertoire is non-functional [7], concluding that, throughout the development process, the sense of smell may have lost importance for primates. It is known that the quantity of functional OR genes is lower in humans compared to other species. Nevertheless, humans have a compassionate sense of smell, which is essential for the discovery of odors that are necessary for maintaining a healthy life, such as the smell of smoke (detection of fire) and the smell of rotten food (to avoid its ingestion) [8].
It is also important to note that the majority of odorants recognized by one species may also be detectable by others, as suggested by Godfrey et al. [9], given that the majority of human OR subfamilies have matching parts in the other species, as is the case of the mouse [9[9][10],10], and other mammals [11].
According to Ngai and co-workers [12], based on their amino acid sequences and phylogenetic distribution, the ORs can be classified as Classes I and II based on the amino acid sequence. Type I ORs bind to hydrophilic odorants, and Class II ORs bind to hydrophobic odorants. The teleost fish, including the goldfish, have only Class I OR genes. On the other hand, the frog and other semiaquatic animals have both Classes I and II OR genes, and initially, it was thought that mammals only contained Class II ORs [13]. These results suggested that the Class I ORs specialized in recognizing water-soluble odorants, and Class II ORs recognize volatile odorants [6]. Studies of the genome sequences have shown many Class I ORs in the human genome [14] and other mammals [11]. Indeed, 10% to 20% of the ORs in mammals are Class I ORs [10[10][11][14],11,14], indicating that Class I ORs are important to mammalian olfaction skills.
The extraction of volatile and non-volatile compounds from plants and animals to develop aromas and fragrances has been carried out since ancient civilizations. It is widely used in cosmetic, pharmaceutical, and food product formulations. Nowadays, the flavors used as additives in beverages and foods are produced via chemical synthesis or extraction [15,16][15][16]. The chemical synthesis of fragrances should be avoided because this is not eco-friendly [17]. Indeed, these extraction processes lead to the formation of undesirable products on the one hand and the growing refusal of consumers to use chemicals added to food and other consumer goods. Nowadays, significant developments are observed in the methodologies for obtaining aromatic compounds, such as microbial and enzymatic biotransformations, de novo synthesis, and genetic engineering tools [16,17,18][16][17][18]. Therefore, natural and sustainable processes are in growing demand from consumers, leading to the opportunity for bio-production alternatives, so novel techniques and ideas are emerging. For instance, (i) plant-cell, tissue, and organ cultures (PCTOC) are an alternative for the production of high molecular weight flavoring molecules, as well as food and beverage additives [19]; (ii) metabolically engineered microorganisms and enzymatic biocatalysis are striking biotechnological options for flavor production [19]. C13-apocarotenoids, derived from the oxidative cleavage of carotenoids, are volatile compounds that contribute to the aromas of diverse flowers and fruits. They are used in the flavor industry and are chemically synthesized. Still, they can be synthesized by bioprocess engineering [20], particularly by (iii) in situ product removal (ISPR), considered a necessary technology for the development of industrial-scale bioprocesses [21]. According to Sá and co-workers [22], enzyme-catalyzed reactions are the most cost-effective strategy from the industrial point of view to reach final green products.
Microbial cell factories, such as Saccharomyces cerevisiae, can be engineered using synthetic biological tools to express synthetic pathways for manufacturing food and beverage flavors. Here, biology is linked to mathematics and computer science. Systems biology, applying computational tools and mathematical models, comprehends complex biological networks and guides synthetic biology design [23].
2. Functional Characterisation and Metabolic Engineering of Flavour Compounds Biosynthesis in Plants
Several hundred volatile compounds have been identified as part of fresh and processed fruits, including fruit juices and vegetables, which cause varied sensory sensations during consumption [25][24] and determine consumer’s choice [26,27,28,29][25][26][27][28]. Even if some fruits present the same compounds known as arenes or aromatics, each fruit has its own fragrance that depends on the aromatic compounds’ composition and concentration [30][29]. Volatile metabolites from fruits and vegetables are lipophilic, represent around 1% of plant secondary metabolites, and are mainly terpenoids, phenylpropanoids/benzenoids, fatty acid derivatives, and amino acid derivatives [27][26]. These compounds are essential for attracting pollinators and seed dispersers and enhancing protection against abiotic and biotic stresses [31][30]. Multiple fruits and vegetables' biochemical pathways are responsible for the volatile compounds’ composition [30][29]. Synthesis of aromatic compounds occurs during fruit ripening and is influenced by several factors, such as temperature and day/night variations [32][31]. Among all volatile compounds, esters are produced by fleshy fruit species during ripening and are the major aroma compounds in apples (Malus domestica), pears (Pyrus communis), and bananas (Musa sapientum). However, volatile esters are in a lower concentration in soft fruits like strawberries (Fragaria × ananassa) [33][32]. Ripeness plays a fundamental role in volatile compounds’ biosynthesis; these compounds are often missing in immature fruits [34][33]. Other factors that can also influence the concentration of the fruit volatile compound are the cultivar, cultural practices, and postharvest management [35][34]. Fruit volatile compounds present in intact tissues are classified as primary metabolites. They can be classified as secondary metabolites if produced due to tissue disruption or fruit injury [36][35]. Therefore, the final fruit aroma profile depends on the sample preparation, whether it is used as an intact fruit, slices, or homogenized fruit sample. According to Valero and Serrano [37][36], volatile compounds determined in entire fruits are related to the consumer smelling perception and the fruit ripening signals, while volatile compounds determined after tissue disruption are more related to the flavor perception during fruit-eating. The biosynthesis of volatile compounds begins from primary metabolic routes, and the formed compounds can be classified as terpenes, fatty acid derivatives, polysaccharide derivatives, and amino acid derivatives [29][28]. According to Croteau and Karp [38][37], the volatile compounds in plants are mainly synthesized from the following pathways: (i) short-chain aldehydes and alcohols (like cis-3-hexenol and n-hexanal) that are synthesized by the action of lipases on lipids, followed by the action of the alcohol dehydrogenases [39][38]; (ii) eugenol, 2-phenyl ethanol, and guaiacol, resulting from the shikimic acid pathway [38][37]; (iii) nor-isoprenoids (like β-ionone and geranylacetone) that result from the degradation of terpenoids [40][39], while two independent pathways can produce terpenoids: the mevalonate pathway in the cytoplasm and the 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway in the plastid [41][40]. The subject of many studies is how to improve the aroma without affecting other fruit or vegetable attributes. Several strategies have been developed to enhance fruit and vegetable-aroma volatile compounds, eliminating or increasing their emission. Those strategies include a wide range of targets, including the metabolic pathways, hormones, transcription factors, and mechanisms involved in the storage or sequestration of volatile precursors [28,42][27][41]. The production of volatile compounds can also be influenced by lower pectin degradation in transgenic fruits by down-regulation of polygalacturonase (PG), pectin methylesterase (PME), and polygalacturonase + pectin methylesterase [28][27]. Metabolic engineering of Aromatic Amino Acids (AAA)-derived pathways is an exciting research topic. It has been used to improve the aroma and flavor of fruits by overexpressing an existing enzyme or blocking a competing path. Manipulation of transcriptional regulation can also be used to change the metabolic profile of AAA-derived volatiles [43][42]. Finally, engineering terpenoids in plants is also a hot topic that includes enhanced disease resistance, the use of allelopathic compounds in weed control, and increased value of ornamentals by producing medicinal compounds [44,45][43][44]. The fundamental factors in terpenoid engineering experiments are the phytotoxicity of the terpenoids introduced, the subcellular localization of both the precursor pool and the raised enzymes, the activity of enzymes that modify the introduced terpenoids and the impacts on other pathways sharing the same precursor pool [44,45][43][44].3. Functional Characterisation and Metabolic Engineering of Flavour Compounds Biosynthesis in Microorganism Cells
Most flavor compounds used in beverages, food, or cosmetics are frequently produced by chemical synthesis, causing a decrease in consumer acceptance due to adding a synthesized chemical compound to the product. Due to health awareness, the flavor companies focused their study on the biological flavor compounds, the so-called “natural flavors” or “bio-flavors” [46,47][45][46]. Plant extracts are a natural source of flavor compounds. Still, their isolation and purification are challenging since the plant materials contain a deficient concentration of the desired compounds, which makes extracting natural flavor compounds expensive, time-consuming, and environmentally unfriendly as it requires relatively large quantities of solvents [48][47]. Production of natural flavor compounds from plant extract also carries severe problems for the environment, ecology, and extent of agricultural soil use. With the growing awareness of environmental sustainability and soil deficiency, microorganisms are engineered for the biosynthesis of natural flavor compounds, linked to advantages such as safety and enough sources of precursors [49][48]. So, a novel promising alternative path for natural flavor synthesis is based on microorganism cell processes, i.e., fermentation or bioconversion of suitable natural precursor using microorganism cells or enzymes (biocatalysis) [44,47,50,51,52][43][46][49][50][51]. Microorganisms present several benefits compared to plants, such as being fast-growing, soil-saving, and controllable. The similar intracellular structure of yeast, especially Saccharomyces cerevisiae, with plant cells led to a widely used host for flavor compounds [53][52]. Two ways to make biotechnologically flavored compounds are de novo synthesis or biotransformation. De novo synthesis is related to having substances using molecules such as sugars. These amino acids, among others, will be metabolized by microorganisms, usually generating a mixture of products in low concentrations [54][53]. Biotransformations are related to single reactions catalyzed enzymatically, so the organism metabolizes the substrate and has a higher potential for producing flavor compounds at a commercial scale [54][53]. Flavor compounds produced by de novo synthesis use the whole metabolism of the microorganism to have a combination of flavor compounds, opposite to biotransformation, where a specific reaction(s) has a significant flavor compound [19]. The development of innovative metabolic engineering and “omic” technologies (genomics, transcriptomics, proteomics, and metabolomics) have started the way for protein and biomolecular pathway engineering. These new methodologies started very significant progress in the metabolic engineering of microorganism cell factories, namely for increased terpenoid synthesis, which may be used to produce flavor compounds at an industrial scale [55][54]. Terpenoids (also called terpenes or isoprenoids) are obtained via the mevalonate biosynthesis pathway or the 2-C-methyl-D-erythritol-4-phosphate pathway, with the former being found in yeast and constitute the two main targets of cell engineering approaches to improve terpenoid production [56,57][55][56]. To produce monoterpenes, which are made directly by terpenoid synthase, Escherichia coli is extensively used for enzyme identification and their significant activity and straightforward expression of these terpenoid synthases in E. coli, as it has a simple genetic manipulation [58,59,60,61,62][57][58][59][60][61]. Yeasts are also becoming attractive, for example, in engineered yeast expressing geraniol synthase [63,64,65,66,67,68][62][63][64][65][66][67]. However, using the yeast cell as an efficient biosynthetic pathway has presented some limitations, such as the biosynthetic pathways are not entirely explained and are associated with the reduced or even missing activity of plant enzymes when expressed in yeast. In addition, decreased cell growth and reduced final product quantity could be related to declines in the native metabolic flux affected by heterogeneous pathways [69,70][68][69]. Also, the cytotoxicity of some natural products could be a limitation to using microorganism cells for producing natural flavor compounds. Identifying new microorganism hosts that can reach more excellent production and developing existing strategies to identify regulatory effects in the central metabolic pathways are fundamental questions about microorganism biosynthesis [46][45]. Several methods and bio-tools to accelerate the microorganism natural products’ biosynthesis in yeast cells have been developed based on omics, metabolic engineering, and protein engineering [71,72,73][70][71][72]. Therefore, strategies to redesign natural biosynthetic pathways are being studied by analyzing the genome and transcriptome data to predict the genes involved in the targeted compound biosynthesis by comparing the transcriptome data between plants with high- and low-production of the target flavor compounds; numerous vital genes could be predicted [72][71]. The low final concentration of the synthesized flavor compounds is frequently related to the reduced enzyme activity on the unnatural substrate, so strategies to improve plant enzyme activity and enhance metabolic flux must be studied [74,75,76][73][74][75]. As the plant natural products' biosynthetic pathways contain many steps, when hosted in the yeast cells, the heterogeneous ways would interact with the native metabolic by competing substrates and co-factors. Therefore, this trouble will limit the targeted flavor compound production by balancing metabolic flux distribution between heterologous pathways and native metabolic shows essential in improving the production of the targeted flavor compound. Also, strategies to decrease toxicity to the hosts are needed [53,77][52][76]. Natural products frequently show cytotoxicity to the microorganism hosts, with the consequence of reducing cell growth and prejudicing the flavor compounds' production. Therefore, several strategies were studied, including two-stage fermentation, pathway separation, and transporters-mediated compound secretion [53,77][52][76]. It is separated into two stages to lessen the adverse effects on cell growth. In the first fermentation stage, the heterogeneous pathway retains calm, and cells grow fast with precursor accumulated; in the second stage, the target pathway is induced to produce the target flavor compounds. Consequently, subcellular separation is a new strategy to decrease product cytotoxicity to the microorganism hosts. An alternative approach to reduce the internal cytotoxicity of natural products is to secrete these compounds external to the cell automatically. To attain this objective, transporters have been considered for transporting the products to the extracellular space. Transporter engineering has been developed to improve transport due to the scarcity of transporters that can transport the flavor natural products [53,77][52][76]. Microorganisms can synthesize flavors as secondary metabolites throughout fermentation (Figure 1) using nutrients such as amino acids and sugars. This ability may be employed as part of food or beverage production processes (some examples include fermented beverages like beer and wine and fermented foods such as vinegar, yogurt, and cheese), which defines the sensory attributes of the resulting product. It can even obtain flavored compounds that label the product as a “natural product.” Solid-state fermentation can provide higher incomes or more suitable product characteristics than submerged fermentation with inferior economic values [78,79][77][78].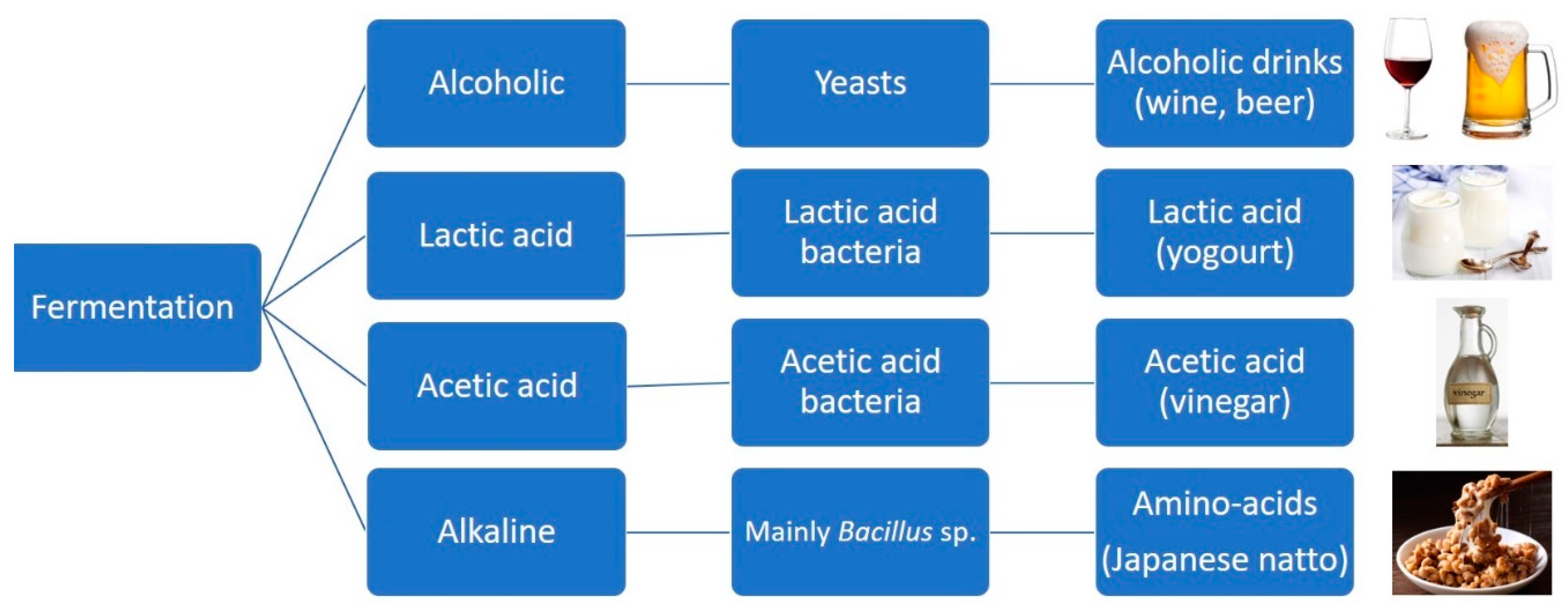
Figure 1. Schematic representation of the common types of fermentation, the microorganisms involved, and the end food products [79][78]. Alkaline-fermented foods constitute a group of less-known food products widely consumed in Southeast Asia and Africa. In alkaline-fermented foods, the protein of the raw materials is broken down into amino acids and peptides; ammonia is released during the fermentation, raising the pH of the final products and giving the food a solid ammoniacal smell. They can be made from different raw ingredients. For instance, Japanese natto is inoculated with a pure culture of Bacillus subtilis var natto [80][79].
4. Mechanisms of Olfaction and Ligand–Receptor Interaction
The peripheral olfactory system was described in 1891 by Santiago Cajal and continues to elude our understanding. In this context, the publication entitled “The Molecular Logic of Smell” by Richard Axel in 1995 [95][80] was significant. It summarises intensively the research done in the late 80s and early 90s, which aimed to study the molecular processes of olfactory translation in the nose´s olfactory epithelium. The olfaction consists of capturing a significant amount of diverse molecular aromas of the natural world, extracting information through personal perceptions related to beverages, flowers, perfumes, and whatever humans encounter daily [96][81]. According to the same authors, olfaction “(…) is a chemosensory processing system that can detect potentially infinite numbers of low molecular-mass compounds, called odorants, which combine at different concentrations, to elicit this complex perception (…)” [96][81]. Thus, the olfactory perception results from ORs' reversible interaction with the odorant molecules (OM) (Figure 2).
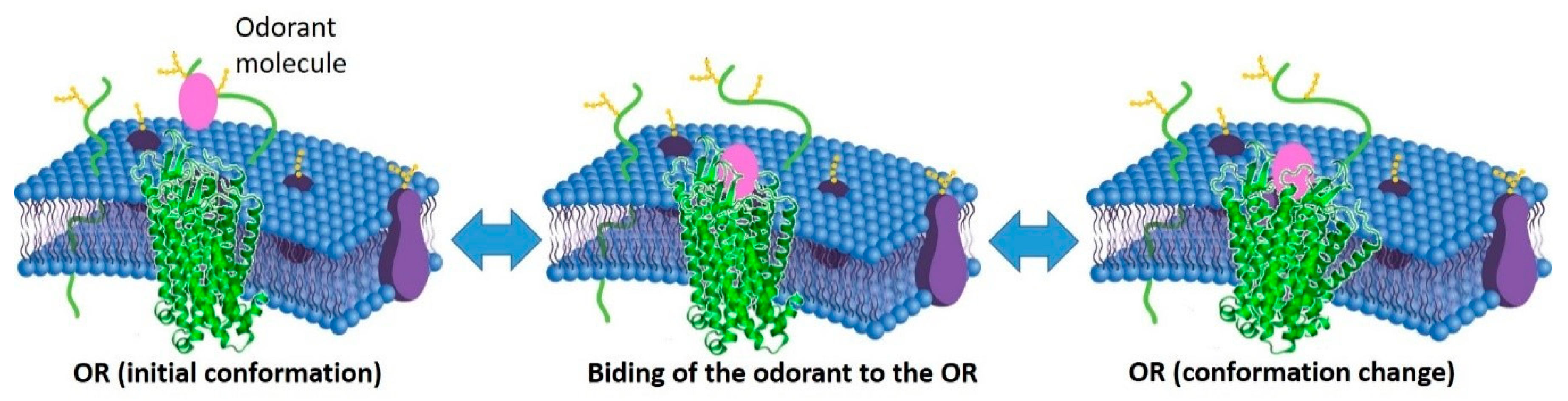

Figure 2. Schematic representation of the interaction between OM (odorant molecule) and the olfactory receptors. This interaction can be analyzed as a sequence of 2 reversible reactions: 1st, the binding of the odorant to the olfactory receptor (OR) and its consequent release, and then, 2nd, the activation (and deactivation) of the OR due to a change of its conformation.
More than one “trillion” (1012) odors can be discriminated [97][82], and some authors say that “The next generation of rich media services will be immersive and multisensory, with olfaction playing a key role (…) for enhancing user quality of experience” [98][83].
In mammals, notably in humans, distinct groups of sensory neurons, which integrate several processing routes, constitute the olfactory system. This system comprises both the central olfactory system and the vomeronasal olfactory system. Each neuron in these systems expresses a type of G-protein-coupled receptor (GPCR) superfamily specialized in detecting a specific type of odor. Thus, the environment presents diverse aromas that can be distinguished by the pattern of all sensory neurons constituting the olfactory system [99][84]. For instance, humans require seven transmembrane G-protein-coupled receptors to identify natural odorants like pheromones [100][85].
But how does the transduction mechanism work? According to Villar and co-workers [101][86], the central olfactory epithelium or olfactory bulb comprises three main cell types: olfactory receptor cells (Figure 3a), sustentacular cells, and basal cells. ORCs (olfactory receptor cells) project a single dendrite to the epithelial surface, where it swells, forming the dendritic knob (Figure 3b).
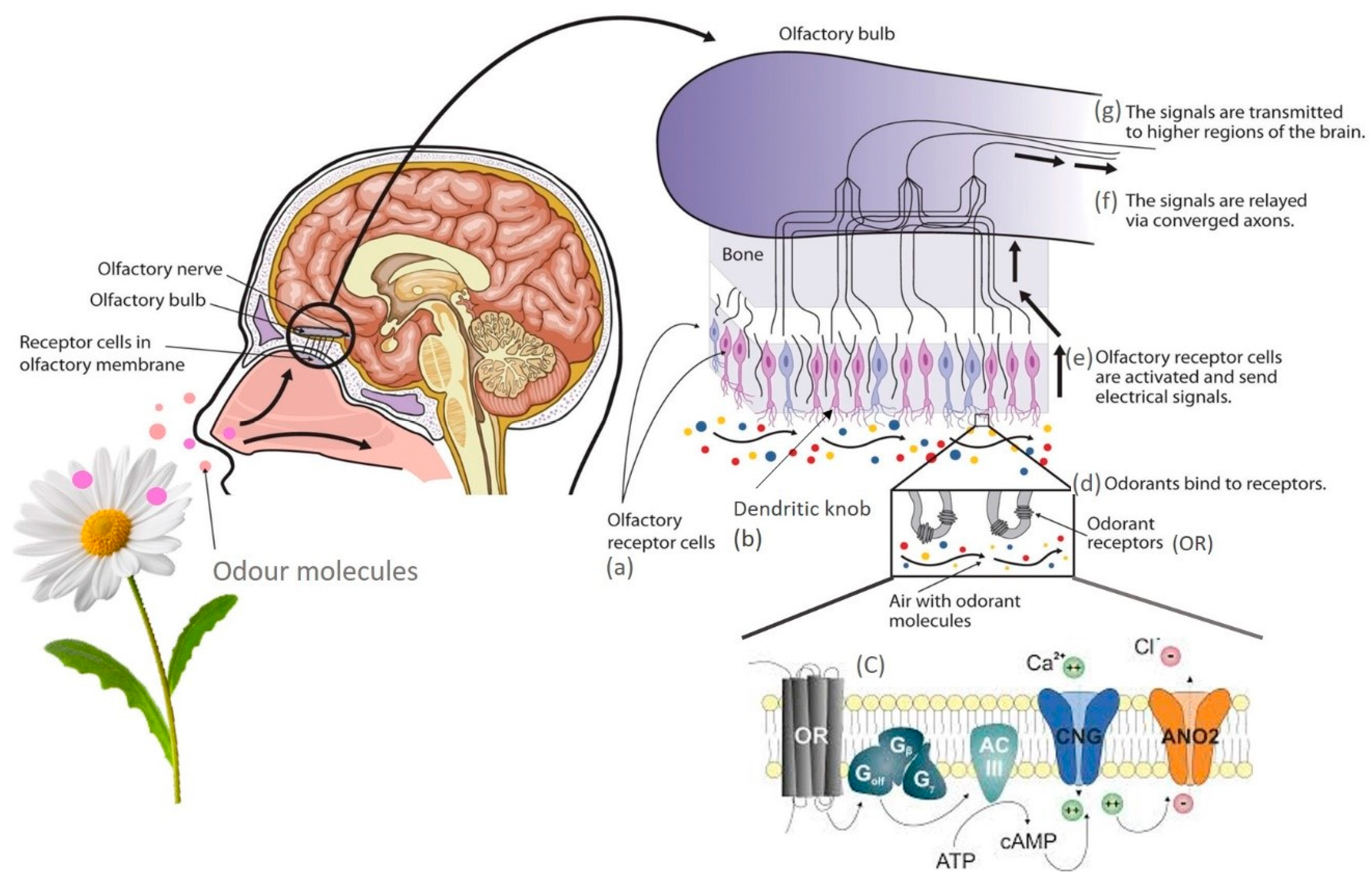

Figure 3. Schematic representation of olfactory transduction mechanism. OR—Odorant receptor protein; Golf—G-protein complex, made up of Gαolf (alpha (α)), Gβ (beta (β)), and Gγ (gamma (γ)) subunits; ACI—Olfactory adenylate cyclase; CNG—Ca2+ Cyclic Nucleotide Gated channel; ANO2—Anoctamin Ca2+ activated Cl− channel; ATP—Adenosine triphosphate; cAMP—Cyclic adenosine monophosphate.
References
- Dunkel, A.; Steinhaus, M.; Kotthoff, M.; Nowak, B.; Krautwurst, D.; Schieberle, P.; Hofmann, T. Nature’s Chemical Signatures in Human Olfaction: A Foodborne Perspective for Future Biotechnology. Angew. Chem. Int. Ed. 2014, 53, 7124–7143.
- Ferrer, I.; Garcia-Esparcia, P.; Carmona, M.; Carro, E.; Aronica, E.; Kovacs, G.G.; Gustincich, S. Olfactory Receptors in Non-Chemosensory Organs: The Nervous System in Health and Disease. Front. Aging Neurosci. 2016, 8, 163.
- Goto, T.; Salpekar, A.; Monk, M. Expression of a testis-specific member of the olfactory receptor gene family in human primordial germ cells. Mol. Hum. Reprod. 2001, 7, 553–558.
- Hillier, L.; Lennon, G.; Becker, M.; Bonaldo, M.F.; Chiapelli, B.; Chissoe, S.; Dietrich, N.; Dubuque, T.; Favello, A.; Gish, W.; et al. Generation and analysis of 280,000 humans expressed sequence tags. Gen. Res. 1996, 6, 807–828.
- Farbiszewski, R.; Kranc, R. Olfactory receptors and the mechanism of odor perception. Pol. Ann. Med. 2013, 20, 51–55.
- Malnic, B.; Gonzalez-Kristeller, D.C.; Gutiyama, L.M. Odorant receptors. The Neurobiology of Olfaction. In Frontiers in Neuroscience; Menini, A., Ed.; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2010.
- Rouquier, S.; Giorgi, D. Olfactory receptor gene repertoires in mammals. Mutat. Res. 2007, 616, 95–102.
- Shepherd, G. The human sense of smell: Are we better than we think? PLoS Biol. 2004, 2, e146.
- Godfrey, P.A.; Malnic, B.; Buck, L.B. The mouse olfactory receptor gene family. Proc. Natl. Acad. Sci. USA 2004, 101, 2156–2161.
- Zhang, X.; Firestein, S. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 2002, 5, 124–133.
- Niimura, Y.; Nei, M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE 2007, 2, e708.
- Ngai, J.; Dowling, M.M.; Buck, L.; Axel, R.; Chess, A. The family of genes encoding odorant receptors in the channel catfish. Cell 1993, 72, 657–666.
- Freitag, J.; Ludwig, G.; Andreini, I.; Rossler, P.; Breer, H. Olfactory receptors in aquatic and terrestrial vertebrates. J. Comp. Physiol. 1998, 183, 635–650.
- Glusman, G.; Yanai, I.; Rubin, I.; Lancet, D. The complete human olfactory subgenome. Gen. Res. 2001, 11, 685–702.
- Shaaban, H.A.; Mahmoud, K.F.; Amin, A.A.; EL Banna, H.A. Application of Biotechnology to the Production of Natural Flavor and Fragrance Chemicals. Res. J. Pharm. Biol. Chem. Sci. 2016, 7, 2670–2717. Available online: https://www.researchgate.net/publication/310460244 (accessed on 1 September 2019).
- Braga, A.; Guerreiro, C.; Belo, I. Generation of Flavors and fragrances through biotransformation and de novo Synthesis. Food Bioproc. Tech. 2018, 11, 2217–2228.
- Gupta, S.; Gupta, C.; Garg, A.P.; Prakash, D. A biotechnological approach to microbial based perfumes and flavours. J. Microbiol. Exp. 2015, 2, 11–18.
- Huccetogullari, D.; Luo, Z.W.; Lee, S.Y. Metabolic engineering of microorganisms for production of aromatic compounds. Microb. Cell Fact. 2019, 18, 41.
- Murthy, H.N.; Georgiev, M.I.; Park, S.-Y.; Dandin, V.S.; Paek, K.-Y. The safety assessment of food ingredients derived from plant cell, tissue and organ cultures: A review. Food Chem. 2015, 176, 426–432.
- Cataldo, V.F.; López, J.; Cárcamo, M.; Agosin, E. Chemical vs. biotechnological synthesis of C13-apocarotenoids: Current methods, applications, and perspectives. Appl. Microbiol. Biotechnol. 2016, 100, 5703–5718.
- Schewe, H.; Mirata, M.A.; Schrader, J. Bioprocess Engineering for Microbial Synthesis and Conversion of Isoprenoids. Adv. Biochem. Eng. Biotechnol. 2015, 148, 251–286.
- Sá, A.G.A.; Meneses, A.C.; Araújo, P.H.H.; Oliveira, D. A review on enzymatic synthesis of aromatic esters used as flavor ingredients for food, cosmetics and pharmaceuticals industries. Trends Food Sci. Technol. 2017, 69, 95–105.
- Fletcher, E.; Krivoruchko, A.; Nielsen, J. Industrial systems biology and its impact on synthetic biology of yeast cell factories. Biotechnol. Bioeng. 2015, 113, 1164–1170.
- Siegmund, B. Biogenesis of aroma compounds: Flavour formation in fruits and vegetables. In Flavour Development, Analysis and Perception in Food and Beverages; Parker, J.K., Elmore, J.S., Methven, L., Eds.; Woodhead Publishing Series in Food Science, Technology and Nutrition: Cambridge, UK, 2015; pp. 127–149.
- Dudareva, D.; Pichersky, E. Metabolic engineering of plant volatiles. Curr. Opin. Biotechnol. 2008, 19, 181–189.
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function, and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32.
- Pech, J.C.; Latché, A.; Van der Rest, B. Genes involved in the biosynthesis of aroma volatiles and biotechnological applications. In Fruit and Vegetable Flavour. Recent Advances and Future Prospects; Brückner, B., Wyllie, S.G., Eds.; Woodhead Publishing Series in Food Science, Technology and Nutrition: Cambridge, UK, 2008; pp. 254–271.
- Aragüez, I.; Valpuesta, V. Metabolic engineering of aroma components in fruits. Biotechnol. J. 2013, 8, 1144–1158.
- El Hadi, M.A.M.; Zhang, F.-J.; Wu, F.-F.; Zhou, C.-H.; Tao, J. Advances in fruit aroma volatile research. Molecules 2013, 18, 8200–8229.
- Pierik, R.; Ballaré, C.L.; Dicke, M. Ecology of plant volatiles: Taking a plant community perspective. Plant Cell Environ. 2015, 37, 1845–1853.
- Belitz, H.-D.; Grosch, W.; Schieberle, P. Food Chemistry, 3rd ed.; Translation from the fifth German edition by M.M. Burghagen; Springer: New York, NY, USA, 2004.
- Beekwilder, J.; Alvarez-Huerta, M.; Neef, E.; Verstappen, F.W.A.; Bouwmeester, H.J.; Aharoni, A. Functional characterization of enzymes forming volatile esters from strawberry and banana. Plant Physiol. 2004, 135, 1865–1878.
- Kader, A.A. A perspective on postharvest horticulture (1978–2003). HortScience 2003, 38, 1004–1008.
- Forney, C.F.; Kalt, W.; Jordan, M.A. The composition of strawberry aroma is influenced by cultivar, maturity, and storage. HortScience 2000, 35, 1022–1026.
- Baietto, M.; Wilson, A.D. Electronic-nose applications for fruit identification, ripeness, and quality grading. Sensors 2015, 15, 899–931.
- Valero, D.; Serrano, M. Postharvest Biology and Technology for Preserving Fruit Quality; CRC Press-Taylor & Francis: Boca Raton, FL, USA, 2010.
- Croteau, R.; Karp, F. Origin of natural odorants. In Perfumes: Art, Science, and Technology; Muller, P.M., Lamparsky, D., Eds.; Elsevier Applied Science: London, UK, 1991; pp. 101–126.
- Galliard, T.; Matthew, J.A. Lipoxygenase-mediated cleavage of fatty acids to carbonyl fragments in tomato fruits. Phytochemistry 1977, 16, 339–343.
- Stevens, M.A. Relationship between polyene-carotene content and volatile compound composition of tomatoes. J. Am. Soc. Hortic. Sci. 1970, 95, 461–464.
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng. Biotechnol. 2015, 148, 63–106.
- Fortes, A.M.; Granell, A.; Pezzotti, M.; Bouzayen, M. Editorial: Molecular and Metabolic Mechanisms Associated with Fleshy Fruit Quality. Front. Plant Sci. 2017, 8, 1236.
- Peled-Zehavi, H.; Oliva, M.; Xie, Q.; Tzin, V.; Oren-Shamir, M.; Aharoni, A.; Galili, G. Metabolic Engineering of the Phenylpropanoid and Its Primary, Precursor Pathway to Enhance the Flavor of Fruits and the Aroma of Flowers. Bioengineering 2015, 2, 204–212.
- Aharoni, A.; Jongsma, M.A.; Kim, T.Y.; Ri, M.B.; Giri, A.P.; Verstappen, F.W.A.; Schwab, W.; Bouwmeester, H.J. Metabolic engineering of terpenoid biosynthesis in plants. Phytochem. Rev. 2006, 5, 49–58.
- Farhi, M.; Marhevka, E.; Masci, T.; Marcos, E.; Eyal, Y.; Ovadis, M.; Abeliovich, H.; Vainstein, A. Harnessing yeast subcellular compartments for the production of plant terpenoids. Metab. Eng. 2011, 13, 474–481.
- Longo, M.A.; Sanroman, M.A. Production of Food Aroma Compounds: Microbial and Enzymatic Methodologies. Food Technol. Biotechnol. 2006, 44, 335–353.
- Vandamme, E.J.; Soetaert, W. Bioflavours and fragrances via fermentation and biocatalysis. J. Chem. Technol. Biotechnol. 2002, 77, 1323–1332.
- Luque de Castro, M.D.; Garcia-Ayuso, L.E. Soxhlet extraction of solid materials: An outdated technique with a promising innovative future. Anal. Chim. Acta 1998, 369, 1–10.
- Nielsen, J.; Keasling, J.D. Engineering cellular metabolism. Cell 2016, 164, 1185–1197.
- Janssens, L.; de Pooter, H.L.; Vandamme, E.J.; Schamp, N.M. Production of flavours by microorganisms. Process Biochem. 1992, 27, 195–215.
- Krings, U.; Berger, R.G. Biotechnological production of flavours and fragrances. Appl. Microbiol. Biotechnol. 1998, 49, 1–8.
- Aguedo, M.; Ly, M.H.; Belo, I.; Teixeira, J.A.; Belin, J.M.; Waché, Y. The use of enzymes and microorganisms for the production of aroma compounds from lipids. Food Technol. Biotechnol. 2004, 42, 327–336.
- Sun, W.; Zhao, Y.-J.; Li, C. De Novo Synthesis of Plant Natural Products in Yeast. In Yeasts in Biotechnology; Thalita, P.B., Ed.; IntechOpen: Rijeka, Croatia, 2019.
- Bicas, J.L.; Molina, G.; Cavalcante Barros, F.F.; Pastore, G.M. White biotechnology for sustainable chemistry. RSC Green Chem. 2015.
- Bution, M.L.; Molina, G.; Abrahão, M.R.E.; Pastore, G.M. Genetic and metabolic engineering of microorganisms for the development of new flavor compounds from terpenic substrates. Crit. Rev. Biotechnol. 2015, 35, 313–325.
- Zwenger, S.; Basu, C. Plant terpenoids: Applications and future potentials. Biotechnol. Mol. Biol. Rev. 2008, 3, 1–7.
- Liao, P.; Hemmerlin, A.; Bach, T.J.; Chye, M.L. The potential of the mevalonate pathway for enhance disoprenoid production. Biotechnol. Adv. 2016, 34, 697–713.
- Albrecht, M.; Misawa, N.; Sandmann, G. Metabolic engineering of the terpenoid biosynthetic pathway of Escherichia coli for production of the carotenoids β-carotene and zeaxanthin. Biotechnol. Lett. 1999, 21, 791–795.
- Kim, S.-W.; Keasling, J.D. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol. Bioeng. 2001, 72, 408–415.
- Leonard, E.; Ajikumar, P.K.; Thayer, K.; Xiao, W.-H.; Mo, J.D.; Tidor, B.; Stephanopoulos, G.; Prather, K.L.J. Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc. Natl. Acad. Sci. USA 2010, 107, 13654–13659.
- Li, Y.F.; Wang, G. Strategies of isoprenoids production in engineered bacteria. J. Appl. Microbiol. 2016, 121, 932–940.
- Ward, V.C.A.; Chatzivasileiou, A.O.; Stephanopoulos, G. Metabolic engineering of Escherichia coli for the production of isoprenoids. FEMS Microbiol. Lett. 2018, 365, 10.
- Paramasivan, K.; Mutturi, S. Progress in terpene synthesis strategies through engineering of Saccharomyces cerevisiae. Crit. Rev. Biotechnol. 2017, 37, 974–989.
- Ignea, C.; Pontini, M.; Maffei, M.E.; Makris, A.M.; Kampranis, S.C. Engineering monoterpene production in yeast using a synthetic dominant negative geranyl diphosphate synthase. ACS Synth. Biol. 2014, 3, 298–306.
- Vickers, C.E.; Williams, T.C.; Peng, B.; Cherry, J. Recent advances in synthetic biology for engineering isoprenoid production in yeast. Curr. Opin. Chem. Biol. 2017, 40, 47–56.
- Wriessnegger, T.; Pichler, H. Yeast metabolic engineering—Targeting sterol metabolism and terpenoid formation. Prog. Lipid. Res. 2013, 52, 277–293.
- Denby, C.M.; Li, R.A.; Vu, V.T.; Costello, Z.; Lin, W.; Chan, L.J.G.; Williams, J.; Donaldson, B.; Bamforth, C.W.; Petzold, C.J.; et al. Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer. Nat. Commun. 2018, 9, 965.
- Wyk, N.V.; Kroukamp, H.; Pretorius, I.S. The Smell of Synthetic Biology: Engineering Strategies for Aroma Compound Production in Yeast. Fermentation 2018, 4, 54.
- Erb, T.J.; Jones, P.R.; Bar-Even, A. Synthetic metabolism: Metabolic engineering meets enzyme design. Curr. Opin. Chem. Biol. 2017, 37, 56–62.
- Smanski, M.J.; Zhou, H.; Claesen, J.; Shen, B.; Fischbach, M.A.; Voigt, C.A. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 2016, 14, 135–149.
- Chemler, J.A.; Koffas, M.A. Metabolic engineering for plant natural product biosynthesis in microbes. Curr Opin. Biotechnol. 2008, 19, 597–605.
- Siddiqui, M.S.; Thodey, K.; Trenchard, I.; Smolke, C.D. Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res. 2012, 12, 144–170.
- Cravens, A.; Payne, J.; Smolke, C.D. Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat. Commun. 2019, 10, 2142.
- Lee, H.; DeLoache, W.C.; Dueber, J.E. Spatial organization of enzymes for metabolic engineering. Metab. Eng. 2012, 14, 242–251.
- Zhao, C.; Gao, X.; Liu, X.; Wang, Y.; Yang, S.; Wang, F.; Re, Y. Enhancing biosynthesis of a ginsenoside precursor by self-assembly of two key enzymes in Pichia pastoris. J. Agric. Food Chem. 2016, 64, 3380–3385.
- Xu, W.; Klumbys, E.; Ang, L.E.; Zhao, H. Emerging molecular biology tools and strategies for engineering natural product biosynthesis. Metab. Eng. Commun. 2020, 10, e00108.
- Istifli, E.S.; Hüsunet, M.T.; Ila, H.B. Cell Division, Cytotoxicity, and the Assays Used in the Detection of Cytotoxicity, Cytotoxicity-Definition, Identification, and Cytotoxic Compounds. Intech. Open 2019.
- Lee, H.-M.; Vo, P.N.L.; Na, D. Advancement of Metabolic Engineering Assisted by Synthetic Biology. Catalysts 2018, 8, 619.
- Anal, A.K. Quality Ingredients and Safety Concerns for Traditional Fermented Foods and Beverages from Asia. A Review. Fermentation 2019, 5, 8.
- Wang, J.; Fung, D.Y.C. Alkaline-Fermented Foods: A Review with Emphasis on Pidan Fermentation. Crit. Rev. Microbiol. 1996, 22, 101–138.
- Axel, R. The molecular logic of smell. Sci. Am. 1995, 273, 154–159.
- Mazzatenta, A.; Cellerino, A.; Origlia, N.; Barloscio, D.; Sartucci, F.; Di Giulio, C.; Domenici, L. Olfactory phenotypic expression unveils human aging. Oncotarget 2016, 7, 19193–19200.
- Bushdid, C.; Magnasco, M.O.; Vosshall, L.B.; Keller, A. Humans can discriminate more than 1 trillion olfactory stimuli. Science 2014, 343, 1370–1372.
- Murray, N.; Lee, B.; Qiao, Y.; Muntean, G.M. Olfaction-Enhanced Multimedia. ACM Comput. Surv. 2016, 48, 1–34.
- Greer, P.L.; Bear, D.M.; Lassance, J.M.; Bloom, M.L.; Tsukahara, T.; Pashkovski, S.L. Family of non-GPCR Chemosensors Defines an Alternative Logic for Mammalian Olfaction. Cell 2016, 165, 1734–1748.
- Rivière, S.; Challet, L.; Fluegge, D.; Spehr, M.; Rodriguez, I. Formyl peptide receptor-like proteins are a novel family of vomeronasal chemosensors. Nature 2009, 459, 574–577.
- Villar, P.S.; Delgado, R.; Vergara, C.; Reyes, J.C.; Bacigalupo, J. Energy Requirements of odor transduction in the chemosensory cilia of olfactory sensory neurons rely on oxidative phosphorylation and glycolytic processing of extracellular glucose. J. Neurosci. 2017, 37, 5736–5743.
More
