Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Camila Xu and Version 1 by Zhangnan Lin.
Butanol fermentation, also known as acetone–butanol–ethanol (ABE) fermentation, refers to the synthesis of butanol by butanol-producing strains using starch crops or sugars as raw materials under strict anaerobic conditions while generating byproducts of acetone and ethanol.
- ABE fermentation
- butanol-producing strains
- low-cost raw materials
1. Introduction
As an important platform chemical, butanol is widely used in industries such as medicine and chemicals, mainly as a solvent or chemical synthesis raw material. At the same time, butanol can be used as a fuel instead of gasoline or as a fuel additive.
With the highlighting of global carbon emissions and environmental issues, butanol production via fermentation has received widespread attention [1,2][1][2]. Butanol fermentation, also known as acetone–butanol–ethanol (ABE) fermentation, refers to the synthesis of butanol by butanol-producing strains using starch crops or sugars as raw materials under strict anaerobic conditions while generating byproducts of acetone and ethanol. ABE fermentation began in the 1850s and has a history of 150 years.
In 1914, Weizmann isolated a strain of butanol-producing clostridia that can synthesize butanol from starch, named Clostridium acetobutylicum (C. acetobutylicum), and its butanol titer can exceed 10 g/L with stable heredity. To date, C. acetobutylicum remains an important species for industrial butanol production, and its discovery laid an important foundation for the industrial development of butanol fermentation.
In the late 1950s, with the rapid development of the petrochemical industry, the cost of synthesizing acetone and butanol using chemical methods became even cheaper, while the high costs of raw materials and subsequent purification of butanol from fermentation resulted in a huge impact on the ABE fermentation industry and many butanol fermentation plants were shut down, leading to a downturn in the butanol fermentation industry [3].
2. Butanol-Producing Strain
Clostridium is the main type of bacterium that is capable of producing butanol, including species such as Clostridium acetobutylicum, Clostridium beijerinckii, Clostridium saccharoperbutylacetonicum, Clostridium saccharobutylicum, Clostridium sporogenes, Clostridium pasteurianum, Clostridium carboxidivorus, Clostridium tetanomorphum, and Clostridium aurantibutyricum [3]. All these strains are strict anaerobic bacteria with rod-shaped cell morphology and are Gram-positive bacteria capable of forming endospores. Table 1 shows the butanol fermentation performance of different strains.Table 1.
Butanol fermentation performance of different strains.
| Strain | Raw Material | Main Product | Butanol Titer (g/L) | Reference |
|---|---|---|---|---|
| Clostridium acetobutylicum | Glucose | Butanol, acetone, ethanol | 10.4 | [4] |
| Clostridium beijerinckii | Glucose | Butanol, isopropyl alcohol | 15.2 | [5] |
| Clostridium saccharoperbutylacetonicum | Glucose | Butanol, acetone, ethanol | 16.2 | [6] |
| Clostridium saccharoperbutylicum | Glucose | Butanol, acetone, ethanol | 9.7 | [7] |
| Clostridium sporogenes | Glucose | Butanol, ethanol, propyl alcohol, isobutanol, methyl butanol | 0.12 | [8] |
| Clostridium perfrigens | Glucose | Butanol, ethanol, propyl alcohol, isobutanol, methyl butanol | 0.02 | [8] |
| Clostridium pasteurianum | Glycerinum | Butanol, 1, 3-propylene glycol | 6.5 | [9] |
| Clostridium carboxidivorus | CO | Butanol, ethanol | 0.37 | [10] |
3. ABE Synthetic Metabolic Pathway of Clostridium
The ABE synthetic metabolic pathway of Clostridium acetobutylicum for the production of butanol is shown in Figure 1 [28]. The major metabolites produced during ABE fermentation include butanol, acetone, and ethanol, accompanied by the generation of CO2 and H2. The entire fermentation process is divided into two stages: the acidogenic phase and the solventogenic phase.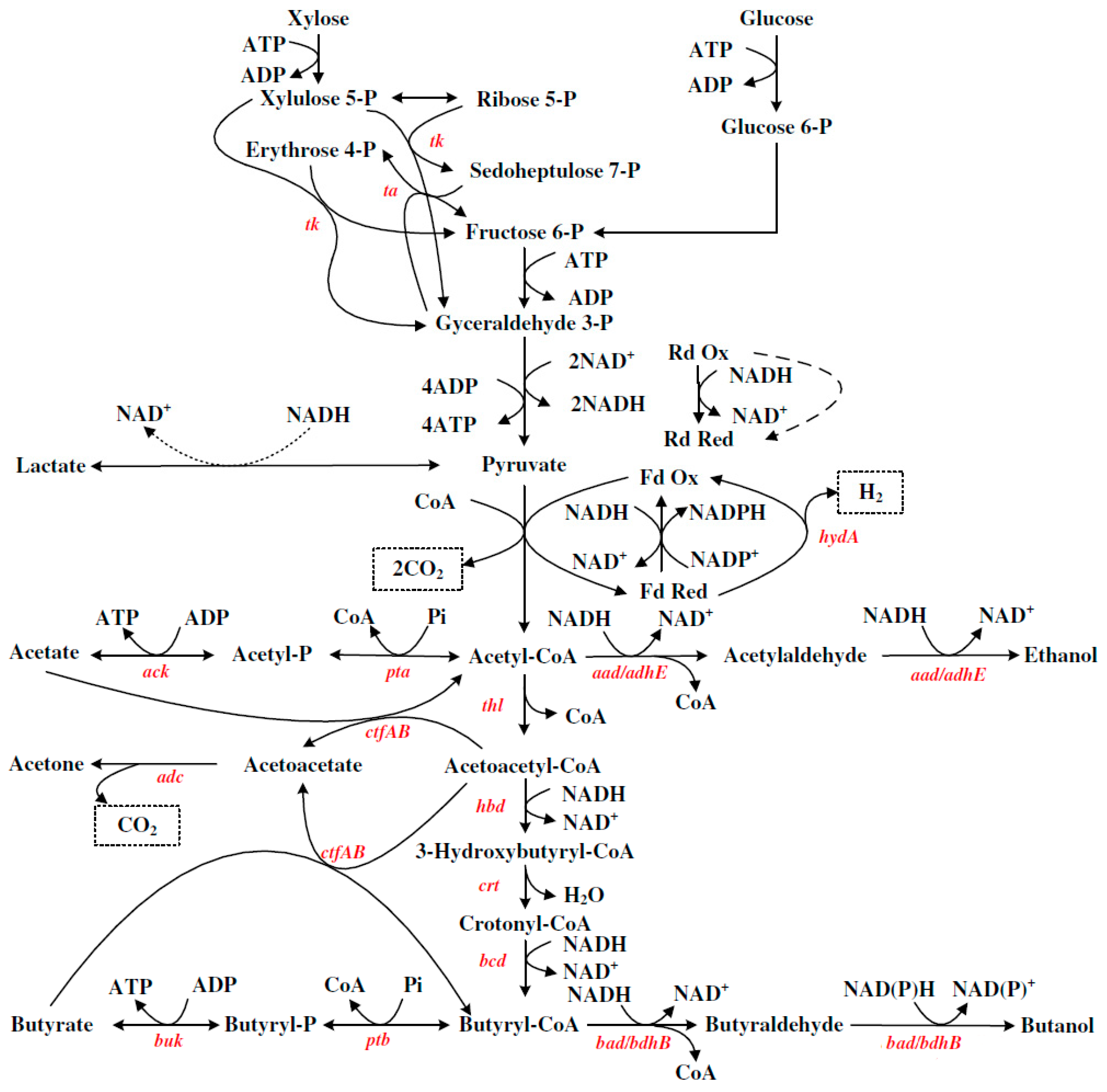
Figure 1. ABE synthesis pathway of Clostridium acetobutylicum [28]. (tk: transketolase; ta: transaldolase; hydA: hydrogenase; aad: acetaldehyde dehydrogenase; adhE: ethanol dehydrogenase; bad: butryraldehyde dehydrogenase; bdhB: butryraldehyde dehydrogenase; buk: butyrate kinase; pta: phosphotransacetylase; ack: acetate kinase; ctfAB: acetate/butyrate:CoA-transferase; hbd: 3-hydroxybutyryl-CoA dehydrogenase; crt: crotonase; bcd: butyryl-CoA dehydrogenase; adc: acetoacetate decarboxylase; ptb: phosphotransbutyrylase).
References
- Algayyim, S.J.M.; Yusaf, T.; Hamza, N.H.; Wandel, A.P.; Fattah, I.M.R.; Laimon, M.; Rahman, S.M.A. Sugarcane Biomass as a Source of Biofuel for Internal Combustion Engines (Ethanol and Acetone-Butanol-Ethanol): A Review of Economic Challenges. Energies 2022, 15, 8644.
- Zhou, Z.; Jing, Y.; Wei, S.; Zhang, Q.; Peng, S.; An, X.; Li, H. Enhancement of butanol production in Clostridium acetobutylicum SE25 through oxidation-reduction potential regulation and analysis of its metabolic mechanisms. Fuel 2022, 331, 125708.
- Moon, H.G.; Jang, Y.S.; Cho, C.; Lee, J.; Binkley, R.; Lee, S.Y. One hundred years of clostridial butanol fermentation. Fems. Microbiol. Lett. 2016, 363, fnw001.
- Huesemann, M.H.; Kuo, L.-J.; Urquhart, L.; Gill, G.A.; Roesijadi, G. Acetone-butanol fermentation of marine macroalgae. Bioresour. Technol. 2012, 108, 305–309.
- Qureshi, N.; Saha, B.C.; Dien, B.; Hector, R.E.; Cotta, M.A. Production of butanol (a biofuel) from agricultural residues: Part I—Use of barley straw hydrolysate. Biomass Bioenergy 2010, 34, 559–565.
- Thang, V.H.; Kanda, K.; Kobayashi, G. Production of Acetone-Butanol-Ethanol (ABE) in Direct Fermentation of Cassava by Clostridium saccharoperbutylacetonicum N1-4. Appl. Biochem. Biotech. 2010, 161, 157–170.
- Berezina, O.V.; Brandt, A.; Yarotsky, S.; Schwarz, W.H.; Zverlov, V.V. Isolation of a new butanol-producing Clostridium strain: High level of hemicellulosic activity and structure of solventogenesis genes of a new Clostridium saccharobutylicum isolate. Syst. Appl. Microbiol. 2009, 32, 449–459.
- Boumba, V.A.; Economou, V.; Kourkoumelis, N.; Gousia, P.; Papadopoulou, C.; Vougiouklakis, T. Microbial ethanol production: Experimental study and multivariate evaluation. Forensic. Sci. Int. 2012, 215, 189–198.
- Ahn, J.H.; Sang, B.I.; Um, Y. Butanol production from thin stillage using Clostridium pasteurianum. Bioresour. Technol. 2011, 102, 4934–4937.
- Lewis, R.S.; Tanner, R.S.; Huhnke, R.L. Indirect or Direct Fermentation of Biomass to Fuel Alcohol. U.S. Patent US20070275447, 29 November 2007.
- Bharathiraja, B.; Jayamuthunagai, J.; Sudharsanaa, T.; Bharghavi, A.; Praveenkumar, R.; Chakravarthy, M.; Yuvaraj, D. Biobutanol—An impending biofuel for future: A review on upstream and downstream processing techniques. Renew. Sust. Energy Rev. 2017, 68, 788–807.
- Pan, H. Study on Extractive Fermentation Conditions and Using Wheat Starch Wastewater to Improve Butanol Production. Master’s Thesis, Jiangnan University, Wuxi, China, 2014.
- Annous, B.A.; Blaschek, H.P. Isolation and characterization of Clostridium acetobutylicum mutants with enhanced amylolytic activity. Appl. Environ. Microb. 1991, 57, 2544–2548.
- Syed, Q.; Nadeem, M.; Nelofer, R. Enhanced Butanol Production by Mutant Strains of Clostridium acetobutylicum in Molasses Medium. Türk. Biyokim. Derg. 2008, 33, 25–30.
- Li, H.-G.; Luo, W.; Gu, Q.-Y.; Wang, Q.; Hu, W.-J.; Yu, X.-B. Acetone, butanol, and ethanol production from cane molasses using Clostridium beijerinckii mutant obtained by combined low-energy ion beam implantation and N-methyl-N-nitro-N-nitrosoguanidine induction. Bioresour. Technol. 2013, 137, 254–260.
- Qureshi, N.; Blaschek, H.P. Recent advances in ABE fermentation: Hyper-butanol producing Clostridium beijerinckii BA101. J. Ind. Microbiol. Biot. 2001, 27, 287–291.
- Jiang, W.; Zhao, J.; Wang, Z.; Yang, S.-T. Stable high-titer n-butanol production from sucrose and sugarcane juice by Clostridium acetobutylicum JB200 in repeated batch fermentations. Bioresour. Technol. 2014, 163, 172–179.
- Jiang, Y.; Xu, C.; Dong, F.; Yang, Y.; Jiang, W.; Yang, S.T. Disruption of the acetoacetate decarboxylase gene in solvent-producing Clostridium acetobutylicum increases the butanol ratio. Metab. Eng. 2009, 11, 284–291.
- Jang, Y.S.; Lee, J.Y.; Lee, J.; Park, J.H.; Im, J.A.; Eo, M.H.; Lee, J.; Lee, S.H.; Song, H.; Cho, J.H. Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum. mBio 2012, 3, 429–493.
- Mann, M.S.; Dragovic, Z.; Schirrmacher, G.; Lutke-Eversloh, T. Over-expression of stress protein-encoding genes helps Clostridium acetobutylicum to rapidly adapt to butanol stress. Biotechnol. Lett. 2012, 34, 1643–1649.
- Scotcher, M.C.; Bennett, G.N. SpoIIE Regulates sporulation but does not directly affect solventogenesis in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 2005, 187, 1930.
- Cong, J. Synergetic Engineering and Metabolic Regulation of Clostridium beijerinckii for Enhancing Xylose Metabolism and Butanol Biosynthesis. Master’s Thesis, Dalian University of Technology, Dalian, China, 2022.
- Abdelaal, A.S.; Yazdani, S.S. Engineering E. coli to synthesize butanol. Biochem. Soc. Trans. 2022, 50, 867–876.
- Liao, H.; Ahmad, K.; Ahsan, M.; Hussain, M.I.; Iqbal, M.W.; Aqeel, S.M.; Hussain, A.; Xia, X. Bacterial metabolic engineering for the production of second-generation (2 G) bioethanol and biobutanol: A review. J. Appl. Microbiol. 2022, 134, lxac061.
- Atsumi, S.; Hanai, T.; Liao, J.C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 2008, 451, 86–99.
- Eric, J.S.; Rossana, C.; Nilu, P.; Myers, S.; Petzold, C.J.; Redding, A.; Ouellet, M.; Keasling, J.D. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb. Cell Fact. 2008, 7, 36.
- Schadeweg, V.; Boles, E. n-Butanol production in Saccharomyces cerevisiae is limited by the availability of coenzyme A and cytosolic acetyl-CoA. Biotechnol. Biofuels 2016, 9, 44.
- Zheng, J.; Tashiro, Y.; Wang, Q.H.; Sonomoto, K. Recent advances to improve fermentative butanol production: Genetic engineering and fermentation technology. J. Biosci. Bioeng. 2015, 119, 1–9.
- Dürre, P. Biobutanol: An attractive biofuel. Biotechnol. J. 2007, 2, 1525–1534.
- Grimmler, C.; Janssen, H.; Krausse, D.; Fischer, R.J.; Bahl, H.; Durre, P.; Liebl, W.; Ehrenreich, A. Genome-wide gene expression analysis of the switch between acidogenesis and solventogenesis in continuous cultures of Clostridium acetobutylicum. J. Mol. Microb. Biotech. 2011, 20, 1–15.
- Lee, J.W.; Na, D.; Park, J.M.; Lee, J.; Choi, S.; Lee, S.Y. Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat. Chem. Biol. 2012, 8, 536–546.
- Jin, X.; Wang, G.; He, B. Research progress and high yield strategy of acetone-butanol fermentation. Chem. Ind. Eng. Prog. 2007, 26, 1727–1732.
More
