TDP-43 gained momentum in the neurodegeneration field when it was first discovered that almost all amyotrophic lateral sclerosis (ALS) cases and as many as half of frontotemporal dementia (FTD) cases present pathological ubiquitinated inclusions of TDP-43. Its involvement in chromatin silencing and nuclear/cytoplasmic shuttling constitute convergent key findings from several biological screens, and several crucial epigenetic factors appear to be able to modify TDP-43-induced degeneration. TDP-43 activity at the chromatin level and its implication in the regulation of DNA transcription and stability -such as DNA repair and regulation of retrotransposons activity- are further supported by a continuously growing amount of studies.
- TDP-43
- TARDBP
- neurodegeneration
- ALS
- FTD/FTLD
- epigenetics
1. Introduction
2. TDP-43 Role in Chromatin Remodeling and Transcription
During the last decade, TDP-43 has been mostly studied for the functions linked to its RNA-binding properties. Notwithstanding this focus, initial studies established the capacity of TDP-43 to bind to ssDNA TG repeats with at least the same efficiency as it does to UG-repeated sequences within RNA [15][16][19][22][15,16,19,22], while binding to other motifs, e.g., to the ssDNA HIV TAR motif from which its name is derived, performed at a slower association rate and at an even slower dissociation rate than it does to (TG)6 stretch [19][20][19,20]. Instead, its ability to bind dsDNA has been documented in different studies [15][20][21][15,20,21] and notably regarding free dsDNA ends [23]. More recently, additional studies have found TDP-43 to be able to specifically bind DNA sequences in promoter regions and affect the expression of several genes [15][19][37][38][39][40][41][42][43][44][45][46][47][48][49][15,19,37,38,39,40,41,42,43,44,45,46,47,48,49], discussed in detail below. Although the function of TDP-43 on chromatin is yet to be fully understood, it is now very clear that the toxic effects of altered TDP-43 can affect chromatin homeostasis.2.1. TDP-43 Is a Global Chromatin Modifier
As already mentioned, the function of TDP-43 as a more general transcriptional activator/repressor has been known for about a decade [40]. Its involvement in chromatin silencing and nuclear/cytoplasmic shuttling constitute convergent key findings from several biological screens, and several crucial epigenetic factors appear to be able to modify TDP-43-induced degeneration [50][51][52][53][50,51,52,53]. For example, using a mosaic genetic screen to study motor neuron degeneration in the Drosophila leg, Sreedharan et al. identified three factors, namely, sgg/GSK3, hat-trick, and xmas-2, needed to mediate TDP-43Q331K toxicity [50]. Interestingly, they noted that the manipulation of these three modifiers did not rescue Wallerian degeneration, another neurodegenerative but TDP-43-independent disease [50]. Among these three proteins, Shaggy (sgg), probably a downstream target of TDP-43, suppressed TDP-43 toxicity without reducing its expression [50]. In parallel, previous screening studies supported a mechanistic link between TDP-43 and Glycogen Synthase Kinase 3 (GSK3). They reported that TDP-43 activates GSK3, while GSK3 inhibition reduces TDP-43 aggregation [54][55][54,55]. Finally, the loss of the two other factors, xmas-2 or hat-trick, implicated in chromatin remodeling and RNA export, affected TDP-43 post-transcriptionally, resulting in a reduction in TDP-43 protein level [50]. In addition to this evidence, two recent studies using the suppressor screen techniques have led to the identifications of other epigenetic modifiers able to contrast TDP-43 neurotoxicity. In these two studies, human TDP-43 was over-expressed in a subset of Drosophila photoreceptor neurons [51], motor neurons, or glial cells [52]. Then, using a combination of shRNA or CRISPR/Cas9 knockdown (KD) screens, these cells were used to identify suppressors of TDP-43 neurotoxicity. Specifically, Azpurua and colleagues used an age-dependent neurobehavioral defect as a primary readout [52], while Berson and colleagues used the red eye degeneration readout [51]. Numerous genes implicated in nucleocytoplasmic transport or in pathways that are deregulated in TDP-43-related neurodegeneration were identified among the glial and motoneuronal TDP-43 suppressors of toxicity. However TDP-43-phenotype suppressors were principally composed of chromatin remodeling and basal transcription machinery factors. In particular, 25% of them were chromatin remodelers. Seven out of eight of these factors promote open chromatin as part of the Trithorax and SWI/SNF (Brahma) complexes, most of them with human known homologues: e(y)3/PHF10; polybromo/BAF180; ash1/ASH1L; enok/KAT6A; br/-; Br140/BRPF1; and mor/BAF170S/MARCC2. The remaining, the Chromodomain-helicase-DNA binding protein 1 (Chd1) with two human homologues, CHD1/CHD2, is an ATPase involved in the remodeling and assembly of chromatin [52]. Further analyses showed that TDP-43 can physically interact with fly Chd1 and human CHD2, impeding their recruitment onto chromatin. Interestingly, both proteins were clearly observed in the chromatin fractions but the Chd1-TDP-43 interaction did not take place on chromatin; rather, it was specifically observed in the cell-soluble fractions both in Drosophila and in human HEK293 cells [55]. By hijacking Chd1, overexpression of TDP-43 resulted in the impairment of correct nucleosome clearing from the gene body of a specific set of stress-protecting genes, preventing their activation [51]. The Chd1-TDP-43 interaction axis might therefore be one way by which the upregulation of TDP-43 sensitizes cells to various stress. Importantly, according to the different brain cells investigated, it was also observed that Chd1 KD could cause an opposite effect on TDP-43 overexpression-mediated toxicity; in particular, instead of counteracting the effects of TDP-43 overexpression upon motor neurons KD, an exacerbation of toxicity was observed upon glial and photoreceptor neurons KD [51][52][51,52]. These data highlight the importance of the cellular context for mediating TDP-43 activity, an important parameter that has been recently observed for TDP-43 pre-mRNA splicing regulatory properties as well [56]. Changing the model organism to C. elegans, it was found that the TDP-43 homolog, TDP-1, could regulate the chromatin localization of another chromatin remodeler, HPL-2, the heterochromatin protein 1 homolog [53]. Direct interaction was found to occur between these two proteins, both in the presence and absence of RNA. In this study, it was shown that TDP-1 facilitates HPL-2 association with active genes to maintain mRNA abundance. In addition, chromatin immunoprecipitation (ChIP) experiments indicated that TDP-1 is present at most of the HPL-2 peaks on chromatin in this organism. Specifically, loss of TDP-1 decreased most of the HPL-2 peaks where (AC)n and (AG)n binding motifs were present. These regions were located predominantly in intronic regions (71%) and promoters (20%) with levels of corresponding RNA decreasing in a tdp-1 mutant worm [53]. As a side note, the intronic localization of TDP-1 on DNA could be related to the propensity of TDP-1/TDP-43 orthologs to bind pre-mRNA chiefly within introns, as previously demonstrated in multiple organisms [34][35][34,35]. At the genome-wide level instead, TDP-43 was found to enrich particularly at promoter regulatory regions, will review in the next sections. Additional evidence of TDP-43 function in chromatin remodeling and its relevance to neurodegenerative diseases comes from the study of nBAFs proteins in cultured mouse motor neurons expressing ALS-linked mutant (G418C and A315T) human TDP-43 [57]. The Brahma-related gene 1 (Brg1)-associated factor (nBAF) chromatin-remodeling complex is critical for neuronal differentiation, dendritic extension, and synaptic function. In this study, the authors showed that nBAF subunits were lost in cultured mouse motor neurons expressing both mutants of human TDP-43. The decrease in nuclear Brg1, BAF53b, and CREST was observed when either mutant was expressed, but also when WT human TDP-43 protein expression was shifted to neuronal cytoplasmic inclusions, thus suggesting TDP-43 as a positive regulator of nBAF expression. In agreement with this conclusion, when co-expressed with mutant TDP-43, the presence of Brg1 delayed the induced dendritic attrition [57]. These data indicate that nuclear loss of TDP-43 can lead to a decrease in nBAF subunits production, either because of a transcriptional repression mechanism or following a defect in RNA processing, potentially leading to RNA nuclear retention, such as the one observed for Brg1 mRNA [57]. Nonetheless, it was interesting to observe that the depletion of nBAF subunits and the delayed attrition upon Brg1 co-expression were not unique to TDP-43; indeed, they were observed also for ALS-linked FUS mutants, and loss of nBAF subunits has also been reported to occur in spinal motor neurons of familial ALS (fALS) and sporadic ALS (sALS) patients with C9orf72 GC expansion (C9ALS) or sALS without mutations in common ALS-linked genes [57]. The contribution of TDP-43, and especially of its ALS-related mutants to more global epigenome alteration, was also recently tested in the human neuroblastoma SH-SY5Y cell line, together with other ALS-causative proteins, SOD1 and FUS [58]. In this work, the authors investigated four modifications on histone H3 tail associated with either transcriptional activation: (i) H3 serine 10 phosphorylation and lysine 14 acetylation (H3S10Ph-K14Ac); and (ii) H3 lysine 4 dimethylation (H3K4me2); or with transcriptional repression marks: (iii) H3 trimethylation of K9H3K9me3; and (iv) DNA methylation. Recombinant adenoviral expression of WT or ALS-related mutants of either TDP-43, SOD1 or FUS proteins all triggered a dose-dependent decrease in cell vitality. However, statistically significant differences in epigenetic marks were limited and specific to the TDP-43 genotype. In particular, a significant decrease in global H3S10Ph-K14Ac was observed for TDP-43M337V, whereas TDP-43WT overexpression led to a significant increase in H3K9me3. On the contrary, no relevant global losses or gains of these epigenetic marks were observed for the TDP-43A382T mutant [58]. In line with these findings, the fly Chd1-TDP-43 interaction study previously mentioned was part of a broad in vivo RNAi screen to search for TDP-43 toxicity modifiers [51]. This screen investigated a total of 84 genes related to various aspects of chromatin biology, including histone methyltransferases (HMTs), demethylases (HDMs), acetyltransferases (HATs), and deacetylases (HDACs), as well as associated factors, histones, and chromatin remodelers. [51]. In addition to Chd1, it allowed for the identification of an additional 4 ‘‘strong’’ and 27 ‘‘mild’’ modifiers, both enhancers and suppressors of TDP-43-mediated eye degeneration. Most of them converged on the conclusion that the TDP-43-mediated toxicity is associated to H3K4me3-linked aberrantly closed chromatin. The modulation of genes that alter other histone methylation marks (repressive H3K27, active-gene body H3K36, or H3K79) mostly had no effect on TDP-43 toxicity [51], with the exception of H3K9 HMT Su(var)3-9. The suppression in flies of Su(var)3-9—that is, the homolog of human SUV39H1—diminished TDP-43-induced toxicity [51]. These observations support the finding by Masala et al. of an aberrant increase in H3K9me3 modification upon ectopic TDP-43WT expression [58]. Note that this effect was not reported for the suppression of G9a, the other well-known H3K9 HMT [51]. Finally, two HDACs, HDAC1 and HDAC6, have been shown to influence and to be influenced by TDP-43, respectively. Thus, it has been shown that the silencing of both HDAC1 in SH-SY5Y and its fly ortholog Rpd3 in Drosophila is able to mitigate the toxic effect induced by TDP-43 expression [48]. Notably, this effect is possibly due to a direct modification of TDP-43 acetylation and consequent cellular localization and functional modulations, notably upon stress (see Section 2.2: TDP-43 and Local/Specific Gene Transcriptional Regulation). In 2010, two peer-reviewed studies showed that TDP-43 was able to bind HDAC6 mRNA, regulating both its mRNA and protein expression in neuronal and non-neuronal cell lines [59][60][59,60]. In one of these studies, Tibbetts’s group demonstrated that this interaction was also mediated by FUS/TLS, which was able to form protein complexes and to share overlapping HDAC6 binding sites with TDP-43 [60]. Conversely, HDAC6 was later shown to exert a deacetylation activity on TDP-43. Indeed, HDAC6 mediated the removal of TDP-43 acetylation at the residues Lys-145 and Lys-192, induced by the CPB acetyltransferase. This was found to decrease the cytoplasmic TDP-43 accumulation in otherwise normal cellular conditions [61]. On the contrary, the formation of TDP-43 aggregates that was induced in case of strong oxidative stress promoted by arsenite could not be deacetylated by HDAC6 despite its interaction with TDP-43, which, overall, contributed to the accumulation of mature aggregates of TDP-43 [61]. In 2020, the relationship between TDP-43 and HDAC6 was further analyzed by Lee and collaborators [62]. They found that the overexpression of HDAC6 in a Drosophila model of TDP-43 proteinopathy reduced the amount of insoluble poly-ubiquitinated proteins and ameliorated the lifespan and climbing defects associated with the overexpression of both TDP-43 and Ataxin-2 (ATXN2). These results indicated that HDAC6 could modulate, albeit in a non-enzymatic manner, the TDP-43 activity via the autophagy–lysosome pathway (ALP) [62]. At the level of gene expression, substantial alterations were observed in the cortices of transgenic mice expressing inducible WT or mutant hTDP-43 lacking the nuclear localization signal (tTA/TDPΔNLS). These alterations appeared even before the onset of significant gliosis and neuronal cell loss [63]. Despite both human TDP-43 transgenes downregulating the endogenous mTDP-43 (by the well-known phenomenon of TDP-43 autoregulation (see specific section)), the mutant lacking the nuclear localization signal showed the most profound changes in gene expression. Among the many processes that were altered in these mice, “DNA–protein complex assembly” pathway was particularly affected and harbored genes coding for major nucleosome proteins. Specifically, many histone variants (H2bp, H3d, H4a/H4b/H4c, and H4h) and several nucleosome assembly protein-1-like1 (NAP1L1) genes were found. While the histone variants were all upregulated, the NAP1L1 genes, on the contrary, were all downregulated [63]. Although these data were obtained using microarray, further RNA-seq analyses on the same model confirmed the alteration in transcription-related pathways and histone transcript levels [64]. In particular, it was observed that Med20, an essential component of the transcription-regulating Mediator complex, and Usp49, a histone H2B deubiquitinase which regulates splicing, were differentially spliced. In parallel, the canonical Histone Hist1h3 and Hist1h4 mRNAs were aberrantly polyadenylated, while at least 10 out of 15 variant histones were slightly but significantly downregulated in the TDPΔNLS bigenic mice [64]. In particular, enhanced cytoplasmic expression of TDP-43 downregulated histone 3′ UTR processing genes, notably Snrpe and Snrpd3, and a similar trend was observed for Lsm1l [64], thus further sustaining a role for TDP-43 in histone transcripts regulation. To relate these findings to the human pathological condition, it is now known that not all cells in the brain of a patient present a reduced load of nuclear TDP-43, and the transcriptome of these cellular populations was recently investigated [65]. To achieve this, Liu et al. successfully separated diseased neuronal nuclei without TDP-43 from nuclei retaining nuclear TDP-43 in a post-mortem FTD and FTD–ALS human brain by combining subcellular fractionation and fluorescent-activated cell sorting (FACS) [65]. Subsequent transcriptome analysis has revealed abundant changes in gene expression associated with loss of TDP-43. In keeping with results obtained from the various animal models, the data from this human material confirmed that many altered genes were involved in histone processing. Furthermore, DNA damage and repair genes were found enriched in addition to genes affecting proteostasis, RNA processing, and nucleocytoplasmic transport. In particular, it was noted that a cluster of 10 altered genes, namely, HUWE1, YY1, MORF4L2, HMGN1, PRKDC, UIMC1, POLB, SFPQ, MSH3, and XRCC5/Ku70, were part of a DNA repair module [65]. DNA methylation is another major epigenetic modification, acting on DNA itself, rather than on the chromatin or nucleosomal proteins wrapped around it. At the biological level, DNA methylation is established via DNA methyltransferases (DNMTs) and is passively erased during DNA replication or, as can be more relevant for neuronal cells, by active replication-independent mechanisms involving oxidations steps mediated by the ten-eleven translocation (TET) enzymes and base excision repair [66][67][68][66,67,68]. DNA methylation in mammals mostly takes place at cytosines (5mC) in the cytosine–guanine dinucleotide context (CpG), but 5mCpH (CpA, CpT, CpC) are also found in the adult mammalian brain [69]. The majority of the CpG are methylated in mammals, with dense CpG islands often unmethylated. CpG islands generally lie in the genes’ regulatory regions and impact transcription. CpG methylation generally has a repressive function, notably controlling promoter activation, but it can also regulate splicing and DNA stability [66][67][70][71][66,67,70,71]. On the other hand, its first oxidized state, the hydroxymethylated C (5hmC), positively influences gene expression, notably in the human brain [72]. No relevant changes in global DNA methylation were observed by Masala et al. in the human neuroblastoma SH-SY5Y cell line overexpressing WT or mutant ALS-linked proteins, including TDP-43, as cited above [58]; however, the brains of ALS patients show a different trend; in fact, altered DNA methylation has been recently observed to occur in human post-mortem CNS tissues from ALS patients using immunohistochemistry. It consisted of higher levels of 5mC and h5mC in the residual lower motor neurons of both sALS and C9ALS compared to the same region in controls [73]. A significantly lower number of neurons with detectable 5mC (mean about 28% vs. >73%) and 5hmC (mean about 51% vs. >87%) was found among neuronal subpopulations with pathological nuclear TDP-43 loss (10% of neurons) compared to those with normal nuclear TDP-43, therefore linking TPD-43 nuclear loss to loss of DNA methylation (despite the direction of causation remaining unknown). Overall, these findings could be connected to differential DNA methylation of several hundreds of genes in ALS spinal cord motor neurons, mostly involved in RNA processing and splicing [73]. Very recently, Catanese and colleagues used multi-omics and machine learning to question the transcriptional, epigenetic, and mutational aspects of heterogeneous human IPSCs-derived motor neurons holding mutants of either C9orf72, TARDBP, SOD1, or FUS, as well as datasets from patients’ biopsies [74]. Analysis of both transcriptome and methylation data resulted in different patterns characterizing the different ALS mutations. Thus, several thousands of DMRs were identified in the ALS sub-group as compared to control, yet a fraction (123 hypermethylated, 179 hypomethylated DMRs) was common to all subgroups, and partially overlapped with the TARDBP mutations (G298S and N390D)-holding subgroup [74]. These results also highlight a deep heterogeneity within the different ALS subtypes on the epigenetic level. Analysis of the DMR-related biological processes, however, indicated that epigenetic abnormalities among ALS iPSCs MNs all contribute to the synaptic alterations (downregulations) observed in all the related transcriptomes, although different sets of synaptic genes were hinted depending on the ALS-related mutation. Nonetheless, all the ALS iPCS-derived MNs displayed upregulation of acetylcholine receptor-binding genes in conjunction with a hypo-methylation of their promoters, notably LY6E, LY6H, and PSCA [74]. In addition, proteomic analysis of proteins co-purifying with TDP-43 in mice brain nuclear extracts has previously identified methyl CpG-binding protein 2 (MeCP2) as an interactor of TDP-43 [75]. MeCP2 is a protein whose defects are responsible for the degenerative Rett Syndrome pathology that binds mC and hmC not only in the CpG context. Interestingly, MeCP2 appears to be implicated in several regulatory contexts similar to TDP-43 (genes and TE transcription and RNA splicing, chromatin loop organization, and heterochromatin structure) [76]. Cell cycle alterations have also been reported following TDP-43 suppression. In two recent publications, TDP-43 activity was linked to sister chromatid cohesion through the splicing regulation of a cohesin complex subunit, namely, Stromal Antigen 2 (STAG2). In particular, depletion of TDP-43 in HeLa and neuroblastoma cell lines upregulated STAG2 exon 30b inclusion [77][78][77,78]. According to those data, cell accumulation was observed in G2/S phase, further supporting the role of TDP-43 in multiple processes involving genome remodeling. Finally, genes related to transcriptional machinery constitute another broad category of TDP-43-phenotype suppressors that have been identified thanks to several screening techniques. These genes include the transcription elongation factor, Su(Tpl) [52][79][52,79], which aberrantly expresses small nucleolar RNAs in TDP-43 pathology [79], TAF1, and e(y)1 orthologs of the mammalian TAF1 and TAF9 transcription factors, members of the TFIID initiation complex, and also Tombola involved in the transcriptional activation of the male germline during meiosis [79]. Specifically, no less than eight genes coding for subunits of the Mediator (Med) complex, mediating RNA-polymerase interaction with transcription factors, were identified by Azpurua et al. [52]. The alteration of another Med subunit, Med20, was identified in mice cortices upon TPD-43 manipulations [64], thus reinforcing a potential role for TDP-43 in gene transcriptional regulation on chromatin. Taken together, these studies on chromatin factors interacting with, modified by, or phenotypically rescuing TDP-43, indicate a potentially important role of TDP-43 as an epigenetic regulator with a high capacity for modulating chromatin, transcriptional processes, and DNA damage/repair pathways. A synthesis of the identified factors can be found in Table 1.| Gene Symbol | Name | Mammalian Orthologs *, Alias | Gene Ontology Function | Contribution to Chromatin Structure/Transcription |
Toxicity Modulation upon Knockdown |
Refs. |
|---|---|---|---|---|---|---|
| Chd1 † | Chromodomain-helicase-DNA binding protein 1 | CHD1, CHD2 | Chromatin remodeling | SILENCING | suppresses/ameliorates | [52] |
| e(y)3 † | Enhancer of yellow 3 | PHF10 | Chromatin remodeling | OPENING | suppresses/ameliorates | [52] |
| polybromo † | Polybromo | BAF180 | Chromatin remodeling | OPENING | suppresses/ameliorates | [52] |
| ash1 † | Absent, small, or homeotic discs 1 | ASH1L | Chromatin remodeling | OPENING | suppresses/ameliorates | [52] |
| enok † | Enoki mushroom | KAT6A | Chromatin remodeling | OPENING | suppresses/ameliorates | [52] |
| br † | Broad | - | Chromatin remodeling | OPENING | suppresses/ameliorates | [52] |
| Br140 † | Bromodomain-containing protein, 140kD | BRPF1 | Chromatin remodeling | OPENING | suppresses/ameliorates | [52] |
| mor † | Moira | BAF170S/MARCC2 | Chromatin remodeling | OPENING | suppresses/ameliorates | [52] |
| MED8 † | Mediator complex subunit 8 | MED8 | Mediator complex | TF–RNA Pol interaction | suppresses/ameliorates | [52] |
| MED11 † | Mediator complex subunit 11 | MED11 | Mediator complex | TF–RNA Pol interaction | suppresses/ameliorates | [52] |
| MED15 † | Mediator complex subunit 15 | MED15 | Mediator complex | TF–RNA Pol interaction | suppresses/ameliorates | [52] |
| MED21 † | Mediator complex subunit 21 | MED21 | Mediator complex | TF–RNA Pol interaction | suppresses/ameliorates | [52] |
| MED22 † | Mediator complex subunit 22 | MED22 | Mediator complex | TF–RNA Pol interaction | suppresses/ameliorates | [52] |
| MED27 † | Mediator complex subunit 27 | MED27 | Mediator complex | TF–RNA Pol interaction | suppresses/ameliorates | [52] |
| MED28 † | Mediator complex subunit 28 | MED28 | Mediator complex | TF–RNA Pol interaction | suppresses/ameliorates | [52] |
| MED31 † | Mediator complex subunit 31 | MED31 | Mediator complex | TF–RNA Pol interaction | suppresses/ameliorates | [52] |
| e(y)1 † | Enhancer of yellow 1 | TAF9 | Transcription Factor | Transcription | suppresses/ameliorates | [52] |
| TAF1 † | TBP-associated factor 1 | TAF1 | Transcription Factor | Transcription | suppresses/ameliorates | [52] |
| Tomb † | tombola | LIN54/TESMIN ** | Male meiosis | Transcription | suppresses/ameliorates | [52] |
| Su(Tpl) † | Suppressor of Triplolethal | ELL, ELL2 | Elongator | Transcription | suppresses/ameliorates | [52] |
| SF2 † | Splicing factor 2 | SRSF1 | Splicing | Transcription | suppresses/ameliorates | [52] |
| Hsp70B (Ba/Bb/Bbb/Bc) † | Heat-shock-protein-70B (Ba/Bb/Bbb/Bc) | HSPA1A | Heat shock Factor | Transcription | suppresses/ameliorates | [52] |
| Chd1 † | Chromodomain-helicase-DNA-binding protein 1 | CHD1 | Chromatin remodeling | TDP-1/TDP-43 impedes Chd1 binding | strongly enhances | [51] |
| dom † | Domino | SRCAP ** | Chromatin remodeling | strongly enhances | [51] | |
| mi-2 † | Mi-2 | CHD3; CHD4; CHD5 ** | Chromatin remodeling | enhances | [51] | |
| kis † | Kismet | CHD6, CHD7, CHD8, CHD9 ** | Chromatin remodeling | enhances | [51] | |
| asf1 † | Anti-silencing factor 1 | ASF1A; ASF1B | Chromatin remodeling | suppresses | [51] | |
| Snr1 † | Snf5-related 1 | SMARCB1 | Chromatin remodeling | suppresses | [51] | |
| mor † | Moira | SMARRC1; SMARCC2 ** | Chromatin remodeling | suppresses | [51] | |
| Dalao † | Brahma associated protein 111kD, Bap111 | SMARCE1 ** | Chromatin remodeling | suppresses | [51] | |
| Chd3 † | Chd3 | CHD3 | Chromatin remodeling | suppresses | [51] | |
| Bap60 † | Brahma associated protein 60kD | SMARCD1 | Chromatin remodeling | strongly suppresses | [51] | |
| MBD-R2 † | MBD-R2 | PHF20; PHF20L1 | Histone Lysine acetyltransferase | OPENING | enhances | [51] |
| Tip60 † | Tat interactive protein 60kDa | KAT5; KAT7; KAT8 ** | Histone Lysine acetyltransferase | OPENING | enhances | [51] |
| CG2051 † | Histone acetyltransferase 1 | HAT1 | Histone Lysine acetyltransferase | OPENING | suppresses | [51] |
| HDAC3 † | Histone deacetylase 3 | HDAC3 | Histone Lysine deacetylase | SILENCING | enhances | [51] |
| Mta1-like † | Metastasis associated 1-like | MTA2; MTA3 | Histone Lysine deacetylase | SILENCING | enhances | [51] |
| Sirt4 † | Sirtuin 4 | SIRT4 | Histone Lysine deacetylase | SILENCING | suppresses | [51] |
| HDAC6 † | Histone deacetylase 6 | HDAC6 | Histone Lysine deacetylase | SILENCING | suppresses * | [51] |
| Scm † | Sex comb on midleg | L3MBTL3; PHC2; SCML1; THAP10 | Transcription Factor | Transcription | suppresses | [51] |
| Spt6 † | Spt6 | SUPT6H | Transcription Factor | Transcription | suppresses | [51] |
| lid † | Little imaginal discs | KDM 4A; 4B; 4D; 4E; 4F; 5A; 5B; 5C; 5D; | Histone Lysine demethylase (H3K4) | SILENCING | strongly suppresses | [51] |
| Utx † | Utx histone demethylase | KDM6A; 6B; UTY | Histone Lysine demethylase (H3K27) | OPENING | suppresses | [51] |
| Asx † | Additional sex combs | ASXL1-3 | Chromatin regulator | SILENCING | suppresses | [51] |
| Ncoa6 † | Nuclear receptor coactivator 6 | NCOA6 | co-activator | Transcription | suppresses | [51] |
| Su(var)3-9 † | Suppressor of variegation 3-9 | SUV39H1 | Histone Lysine methyltransferase (H3K9) | SILENCING | suppresses | [51] |
| ash2 † | Absent, small, or homeotic discs 2 | ASH2L | Histone Lysine methyltransferase (H3K4) | OPENING | enhances | [51] |
| Su(z)2 † | Suppressor of zeste 2 | BMI1; PCGF1; 2; 5; 6; RING1B; RNF2 | Histone Lysine methyltransferase, histone monoubiquitination (H2A-K119) |
OPENING | enhances | [51] |
| Set1 † | SET domain containing 1 | KMT2B | Histone Lysine methyltransferase (H3K4) | OPENING | enhances | [51] |
| Wdr82 † | WD repeat domain 82 | WDR82 | Histone Lysine methyltransferase (H3K4) | OPENING | enhances | [51] |
| HPL-2 †2 | Heterochromatin Protein-like 2 | HPL1 | Chromatin remodeling | TDP-1 bridges HPL-2 to chromatin | n.d. | [53] |
| nBAF complex | Neuronal Brg1/BRM Associated factor | SS18L1, NBAF; CREST; SYT homolog 1 | Neuronal chromatin remodeling complex nBAF component | n.d. | [57] | |
| sgg † | Shaggy | GSK3 | Control of cellular pathways and metabolism | suppresses | [50] | |
| htk † | Hat-trick | Chromatin remodeling | suppresses | [50] | ||
| xmas-2 † | Xmas-2 | Transcription and RNA export | suppresses | [50] | ||
| H3C14 | H3S10Ph-K14Ac: Histone 3 (serine 10 phosphorylation/lysine 14 acetylation) | H3 | Histone tail PTM | Transcriptional activation/repression | n.d. | [58] |
| H3C14 | H3K9me3: Histone 3 (methylation of lysine 9) | H3 | Histone tail PTM | Transcriptional repression | n.d. | [58] |
| HDAC1 | Histone deacetylase 1 | HDAC1 | Histone/Protein Lysine deacetylase | SILENCING *** | n.d. | [48] |
| H2b | Histone cluster 1, H2bp | HIST1H2BP | Nucleosome assembly | Downregulated in the TDPΔNLS | n.d. | [63] |
| H3d | Histone cluster 1, H3d | HIST1H3D | Nucleosome assembly | Downregulated in the TDPΔNLS | n.d. | [63] |
| H4a/H4b/H4c/H4h | Histone cluster 1, H4a, b, c, h | HIST1H4 | Nucleosome assembly | Downregulated in the TDPΔNLS | n.d. | [63] |
| Nap1l1 | Nucleosome assembly protein 1-like1 | NAP1L1 | Nucleosome assembly | Upregulated in the TDPΔNLS | n.d. | [63] |
| Med20 | Mediator Complex subunit 20 | MED20; SRB2; TRFP; PRO0213 | Mediator Complex/transcriptional coactivator | Transcriptional activation | n.d. | [64] |
| Usp49 | Ubiquitin specific processing protease 49 | USP49 | H2B Histone deubiquitinase | Transcriptional activation/regulation of mRNA splicing | n.d. | [64] |
| HUWE1 | HECT, UBA, WWE domain Containing E3 Ubiquitin Protein ligase 1 | HUWE1 | Histone/protein Ubiquitination | n.d. | [65] | |
| YY1 | Yin and Yang 1 transcription Factor | YY-1; INO80S | Transcription factor | Transcriptional activation/repression | n.d. | [65] |
| MORF4L2 | Mortality Factor 4-like 2 | MRGX; KIAA0026; MORF-Related Gene X | Heterochromatin assembly/histone modification | Transcriptional activation | n.d. | [65] |
| HMGN1 | High-Mobility Group Nucleosome Binding Domain 1 | HMG14; MGC104230; FLJ27265 | Chromatin remodeling | n.d. | [65] | |
| PRKDC | Protein Kinase, DNA-Activated, Catalytic subunit | DNA-PKcs; DNPK1; P460; DNAPKc; XRCC7; P350 | DNA repair and recombination | n.d. | [65] | |
| UIMC1 | Ubiquitin Interaction Motif Containing 1 | RAP80; Retinoid X Receptor-Interacting Protein 110 | DNA repair | Ubiquitination/Transcription repression | n.d. | [65] |
| POLB | DNA Pol Beta | DNA Pol Beta | DNA excision and repair | n.d. | [65] | |
| SFPQ | Splicing Factor Proline and Glutamine Rich | PSF; PPP1R140; HPOMp100; POMP100 | Splicing | Enables DNA binding activity | n.d. | [65] |
| MSH3 | MutS Homolog 3 | MRP1; DUP; HMSH3; Mismatch Repair Protein | DNA mismatch repair system (MMR) | Postreplicative DNA mismatch repair | n.d. | [65] |
| XRCC5 | X-Ray Repair Cross Complementing 5 | Ku86; KARP-1; KU80; KUB2 | DNA repair | DNA DSB repair by NHEJ | n.d. | [65] |
| XRCC6 | X-RAY Repair Cross Complementing 6 | KU70; Cells6; G22P1; ML8 | DNA repair | DNA DSB repair by NHEJ | n.d. | [65] |
| STAG2 | Stromal Antigen 2 | SA2; SCC3B; SA-2; Cohesin Subunit SA-2 | Chromatin remodeling | Cohesion of sister chromatids after DNA replication | n.d. | [77][78][77,78] |
| Ell, Su(Tpl) † | Elongation Factor for RNA Pol II | ELL; ELL2; C19orf17; PPP1R68 | Elongator | Transcription regulation | enhances | [52][79][52,79] |
2.2. TDP-43 and Local/Specific Gene Transcriptional Regulation
Since 1995, when TARDBP was first identified as being able to bind TAR motif within HIV proviral DNA [15], a dozen additional studies identified TDP-43 as a potentially important player in the regulation of other specific genes, according to several modalities (Figure 1).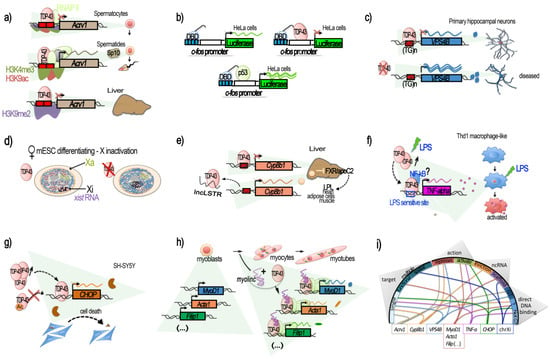
2.3. TDP-43 and Genome-Wide Transcriptional Regulation
Cellular evidence for a potential broad function of TDP-43 in transcriptional regulation in physiological conditions has been supported by confocal and electron microscopy studies combined with in situ detection of transcription [82]. In this work, TDP-43 was found distributed throughout the euchromatin of the primary sensory ganglia neurons of rats and to be enriched at perichromatin fibrils, i.e., mRNA transcription and processing sites. In particular, TDP-43 signal was evident at sites of nascent pre-mRNA. Conversely, only weak TDP-43 immunolabeling was found in nuclear speckles that represent areas enriched by splicing factors. Finally, transcriptionally silent constitutive centromeric and telomeric heterochromatin, as well as Cajal bodies, did not concentrate TDP-43 [82]. These microscopic observations globally concur with chromatin fractionation analyses followed by Western blot experiments performed in HeLa cells [24]. About 7 years later, a higher resolution of the genome localization of TDP-43 was first given on its Drosophila homolog, TBPH, by a plethora of experiments based on ChIP-seq, RNAi depletion, transcription blockade, affinity chromatography, and immunoprecipitation conducted in Drosophila [83]. This study confirmed the presence of TDP-43/TPBH at gene regulatory locations, where TBPH appeared to bind chromosomes at specific sites, and not only at splicing-related features, such as gene bodies, but also at genes, enhancers, and Polycomb response elements (PREs) bound by cohesin. At these regulatory regions and genes, TBPH was found to ensure high levels of Nipped-B and cohesin on the sites [83]. As described in mouse or human, TBPH targeting has been linked to the presence of TG reach repeats in the non-template strand of these genes. Based on the obtained results, a model was proposed forecasting that UG repeats on the nascent transcripts recruit TDPH via RRM1 domain binding, then the Nipped-B and cohesin complex are recruited. In turn, Nipped-B boosts the TDPH presence at poorly-transcribed regulatory regions, such as enhancers and PREs. Continued transcription is not required to maintain their binding once it has been established [83]. In higher organisms, such as human and mouse, PRE are not conserved, and the mechanisms and modalities of Polycomb-repressive complexes, PRC2 or PRC1, loading to chromatin are various and multifactorial and still represent a field of intense research [84][85][86][87][84,85,86,87]. The colocalization or interaction between TDP-43 and PRC2 or PRC1 have not been reported yet, but PRC2 target genes were recently found derepressed in post-mortem brain samples from ALS/FTD patients with C9orf72 (C9) repeat expansions, and the PCR2 HMT subunit EZH2 was found largely insoluble [88]. Interestingly, a deep characterization of the extent of TDP-43 associations with chromatin in the human was recently obtained via the analysis of ENCODE-released ChIP data in HEK293T cells [89]. TDP-43 general genome-wide localization at gene promoters was confirmed. In particular, a strong enrichment was observed at promoters in association with high RNA Pol II presence. However, TDP-43 did not bind, at least directly, RNA Pol II, nor did it string along with it within the gene body of active genes [89]. siRNA-induced silencing of TDP-43 reduced the transcription of thousands of genes, as analyzed by GRO-seq, including protein-coding, antisense non-coding, and lincRNA genes, and conversely, it activated only a little fraction of genes in each of these categories. Several miRNA and snRNA were also affected [89]. Still, there was no compelling evidence of a relationship between TDP-43 abundance at a gene promoter and the degree of transcriptional change after TDP-43 loss. Instead, TDP-43 loss resulted in increased transcription of repetitive elements found within expressed genes belonging to the Alu class of non-autonomous retrotransposons, the affected density of which corresponded to changes in gene transcription [89]. As TDP-43 did not appear to interact with RNA Pol II, nor with the affected Alu at these regions, the mechanisms behind it may be indirect, affecting other TDP-43 pathways. At the beginning of 2021, Maor-norf and colleagues, working on mouse cortical culture and combining ATAC-seq and RNA-seq, investigated the consequences on the chromatin accessibility of ALS-related protein overexpression [90]. Though the main outcome of the publication relies on the C9orf72 poly(PR) ((PR)50) mutant and the interesting finding that (PR)50-induced neuronal death can be dampened via p53 inhibition, the authors nonetheless obtained very interesting findings regarding TDP-43 neuronal overexpression. Despite the neurodegeneration looking “grossly similar”, the modifications of the underlying chromatin and gene expression programs were different. TDP-43 and C9orf72 (PR)50 conveyed unique chromatin and transcriptional footprints. Loss of chromatin accessibility was observed for all lentiviral-treated cultures, even the GFP control; however, a gain in chromatin accessibility was observed for TDP-43 after 60 h and associated with a chromatin more accessible for a variety of TFs, as evidenced via ChromVar, and several gene co-expression networks were deregulated [90], findings that definitively deserve deeper characterization. As a result of these investigations, it appears that TDP-43 has a broad ability to affect the transcription of all categories of genes, from coding to non-coding classes, to a degree that is greater than had been previously appreciated. This can be achieved indirectly through the modulation of genes involved in chromatin remodeling and transcription, or directly via association with genes promoters. While direct, multimodal regulation, either repressive or activating, has, as discussed above, been reported for several genes, understanding of the general function of TDP-43 at the TSS of active genes genome-wide still requires further investigation. It was proposed that the transcription-independent binding of TBPH, and hence possibly of TDP-43, could serve to reduce the fluctuations in the levels of transcription over time, with the intriguing possibility that the aggregate-prone low-complexity C terminal domains in TBPH might also facilitate enhancer–promoter looping or loops stabilization [83]. Remarkably, Nie and colleagues reported very recently on the requirement of the maternal (oocyte) TDP-43 protein to activate the zygotic genome during embryogenesis by promoting RNA Pol II transition from a paused to an elongating state [91]. Experiments driven in mouse showed that maternal TDP-43 proteins translocate from the cytoplasm to the nuclear space from the 2C stage, where they localize at RNA Pol II clusters and associated with RNA Pol II, as shown via a proximity ligation assay (PLA), co-IPs and ChIP-seq-like Stacc-seq technology. TDP-43 also co-occupies with RNA Pol II at the promoters of ZGA genes at the late 2C stage. Importantly, the deletion of maternal TDP-43 led to defective zygote genome activation. Indeed, their results support the fact that TDP-43 promotes the expression of ZGA genes by activating transcription of RNA Pol II elongation from its pausing through RNA Pol II CTD Ser2 Cyclin T1 phosphatase during mouse maternal-to-zygotic transition [91]. However, as observed for Acrv1 gene promoter activity during mouse spermatogenesis [40], the here-described essential role of TDP-43 in early mouse embryogenesis is stage-specific, as its absence at an earlier stage, i.e., in mouse full-grown oocytes, only mildly affects gene expression [91]. Overall, TDP-43 seems to have a pivotal role in the cell fate gene induction of different tissues (spermatogenesis [40], myogenesis [42], and embryogenesis [91]).2.4. TDP-43 and DNA Repair
Alongside the TDP-43 function in gene transcriptional regulation, several recent studies have strengthened the potential role of TDP-43 in genome stability and DNA repair [23][92][93][94][95][96][23,92,93,94,95,96]. Upon DNA damage induced in pluripotent stem cell (iPSCs)-derived motor neurons from a healthy subject, or in differentiated neuronal line SH-SY5Y, normal TDP-43 is rapidly recruited at double-stranded breaks (DSB) sites. TDP-43 was shown to stably interact with DNA damage response (DDR) and neighbor homologous end-joining (NHEJ) repair factors. Specifically, it acted as a scaffold protein for the DDR complex (γH2AX, pATM, Ku70, p53BP1) and the break-sealing XRCC4-DNA ligase 4 complex (XRCC4, lig4, XLF), mediating its recruitment at induced DSB sites [23][93][23,93]. In vitro experiments further showed that TDP-43 can directly bind dsDNA oligonucleotides with free blunted ends, but not if ends are biotin-blocked or in case of a ssDNA break [23], thus supporting the TDP-43 recognition and binding of DSBs in the genome. It is notable that TDP-43 was found pre-complexed with some proteins of the repair machinery, i.e., Ku-70, Ligase 4, and XRCC4, but also with p-53BP1 and γH2AX, already in the absence of artificially induced breaks [23]. Upon DSB induction, these interactions were significantly enhanced [23]. The authors showed that TDP-43 specifically interacted with NHEJ proteins and remained at DSB until repair completion [23]. In SH-SY5Y, TDP-43 Q331K mutant prevented the nuclear translocation of XRCC4-DNA ligase 4, and cells showed elevated levels of reactive oxygen species, thus contributing to both DNA damage production and irresolution [93]. As expected from these findings, deprivation of TDP-43 led to an accumulation of DNA damage. As such, TDP-43 deprivation in cycling iPSCs-derived NPCs and SH-SY5Y conveyed an accumulation of endogenous DSBs, despite DDR activation, with an increase in γH2AX, p53BP1, and pATM at 96 h after TDP-43 KD [23]. Subsequently, cells proceeded to apoptosis [23]. This TDP-43 function is universal among metazoans as TDP-1-lacking worms also have an impaired DSB repair [23]. An increased in yH2AX upon TDP-43 shRNA was also observed in differentiated and not differentiated SH-SY5Y, as well as in NPC-derived motor neurons [23], and in SH-SY5Y cells overexpressing a A382T TDP-43 mutant [97]. However, TDP-43 knockdown from NSC-34 motor neuron-like cells or primary cortical neurons resulted in a significant decrease in both γH2AX foci and global γH2AX amounts [95], thus underlying probable cell-type and assays specificities. Since neurons are post-mitotic cells, they are particularly dependent on the NHEJ DNA repair pathway, unlike other cells that can take support from the less-error-prone DNA repair by homologous recombination (HR) [98]. Two mechanistically distinct NHEJ DNA repair pathways exist: the classical (C-NHEJ) is Ku70, Lig4, and Rad51-dependent [99]; the alternative is NHEJ (alt-EJ), which is not dependent on these factors [99]. From the evidence presented above, and further sustained by an additional study using GFP-reporter systems specific to either of the two NHEJ DSB repair mechanisms, TPD-43 clearly participates in classical NHEJ DSB repair [95]. On the contrary, neither mutant (Q331K, A315T) nor wild-type TDP-43 participate in the modulation of the alt-EJ [95]. TDP-43’s role at DSB sites for NHEJ repair and related dysfunctions in contexts of altered TDP-43, such as ALS, is illustrated in Figure 2A–C.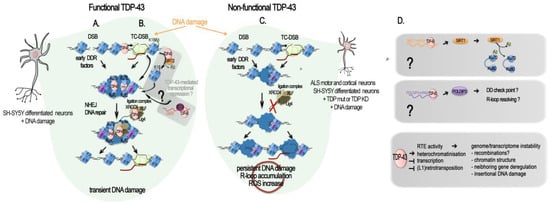
2.5. TDP-43 and Regulation of the Genome Dark Matter
The role of TDP-43 in genome stability might not only be linked to the protection of DSB at active transcription, i.e., at transcribed genes; growing evidence shows that it has a function in maintaining silent the so called “Dark matter” “selfish DNA” of our genomes (Figure 2D and Figure 3A). In their recent work on the post-mortem brain of FTD patients, Liu and colleagues integrated their transcriptome analyses with ATAC-seq to examine changes in chromatin accessibility in TDP-43-negative nuclei relative to TDP-positive nuclei from the same samples for seven FTD and FTD–ALS brains [65]. They identified 3457 significantly differentially accessible genomic regions, the great majority (75.2%) of which corresponded to a more closed chromatin in TDP-43-negative nuclei. However more accessible chromatin was enriched for elements typically found in heterochromatic regions, depleted from classical genes, suggesting a similar overall euchromatinization in the TDP-43 pathologic nuclei similar to the one observed in mutant mice [64][65][64,65]. The ability of TDP-43 to maintain genome dark matter silence is not so surprising given that it was discovered as a transcriptional repressor of the HIV provirus (Figure 3B). Mammal genomes are full of remnants from ancient retroviral infections of the germ line cells that have resulted in the integration of proviral genomes into the DNA of offspring. Over time, some of these integrations led to the fixation of the proviruses in the gene pool of the host population, thus becoming an endogenous retrovirus (ERV). In parallel, their subsequent within-germline propagations by means of retrotranspositions or reinfections (copy–paste-like mechanisms) over millions of years led to the formation of several multicopy families that group under the long terminal repeat (LTR) class of TEs. Retroviral ORFs (gag (viral core proteins), pro/int/pol (enzymatic proteins: protease integrase, reverse transcriptase polymerase), env (envelope glycoprotein), and accessory proteins) accumulated disruptive nonsense mutations and proviruses often recombined leading to solo-LTRs. In the human, HERV-derived copies and fragments represent about 8% of our genomic DNA [113][114][115][116][113,114,115,116]. Insertional and recombinational polymorphism of some HERV copies exists within the population, and they belong to the more recent HERV-K HML-2 family [117][118][119][117,118,119]. Several ORFs from different HERV families still remain, and various examples of domesticated ERV proteins have been reported, especially for env glycoproteins [120][121][122][123][120,121,122,123]. In addition, LTRs contribute greatly to “cellular” gene regulatory sequences such as promoter, enhancer, or polyadenylation signals [116][124][116,124]. In contrast to their human counterparts, some murine endogenous retroviruses (mERV) proviruses still can synthesize infectious particles and retrotranspose. The long interspersed elements (LINEs) constitute another important class of retrotransposons; they are able to conduct autonomous self-propagation via another copy–paste mechanism thanks to their ORFs encoding ribonucleoproteins, endonuclease, and reverse transcriptase. They account for 17% of the human genome, and several of them, albeit representing a small fraction, are polymorphic within the population [125]. They directly contribute to the expansion of short interspersed elements (SINEs), another class of TE, non-autonomous, and of pseudo- and retrogenes in the genome [126][127][126,127]. An additional class of TE, the DNA transposon, mobilizes through a cut–paste mechanism. Together, the TEs occupy nearly 46% of the human genome and 39% of the mouse genome [113][128][113,128]. Contrary to human ERV, retrotransposition-competent (RC) copies of LINES in human and mouse genomes are numerous. A small number of these RC-L1s loci, Hot-L1, are highly active [125], notably in human and mouse developing brains [129][130][129,130], and result in normal brain genome mosaicism [131]. Overall, TE subfamilies are species specific, but they rely on the same molecular mechanisms for their control and propagation [132], notably epigenetic mechanisms, including CpG DNA methylation. Indeed, genomes evolved defenses against their detrimental potential, and TE are generally silenced by DNA methylation and heterochromatin marks such as H3K9me3, acting as major barrier against their activation [133][134][135][136][133,134,135,136]. Thanks to the efforts of recent studies, TDP-43 has recently been shown to play a role in regulating them at several levels. Being highly repetitive by nature, TEs are routinely dismissed from deep-sequencing analyses if not under specific focus, although new sequencing modes have greatly improved their mappability. However, by reanalyzing a series of deep sequencing datasets from RIP-seq and iCLIP-seq from normal brains of rat, mouse, and human, the group of Dubnau uncovered an extensive binding of TDP-43 to TE transcripts [137]. In this way, several ERV/LTR classes, but also SINEs and LINEs and some DNA repeats, were identified. Interestingly, although peaks that map over RefGene (classical “cellular” protein-coding and non-coding genes) annotations were similarly distributed for both FUS and TDP-43 iCLIP-seq experiments in mouse, only TDP-43 clearly targeted TE-transcripts for binding, and via a similar sequence motif (UGUGU), as reported for “cellular” transcripts [137]. Importantly, this physiological binding of TDP-43 to TEs was observed to be altered in at least two of the major TDP-43 proteinopathies: FTLD and in a specific subtype of ALS [137][138][137,138]. In FLTD patients, reanalysis of iCLIP-seq data showed a reduced association between TDP-43 and TE transcripts for all major classes, including SINE, LINE, LTR, and a few DNA transposon elements, principally originating from intergenic locations [137]. This reduction was more pronounced for TE than for “cellular genes”. Furthermore, TDP-43 depletion and overexpression (acting as a dominant-negative) in mouse brain both conveyed the robust overexpression of some tens to several hundreds of copies of TE-derived transcripts; with the vast majority of them corresponding to those identified in the iCLIP-seq data [137]. With regard to ALS, the dominant feature characterizing a specific sub-group consisting of 20% of ALS patients was a marked retrotransposons re-activation [138]. This recent study applied machine learning-assisted analysis of RNA-seq from frontal and/or motor cortex samples of a cohort of ALS patients and controls [138]. Again, this ALS-TE subgroup included TEs from the LINE, SINE, and LTR classes, as well as several individual retrotransposons from the HERV-H, LINE L1M2a, and L1PA6, and SINE–VNTR–Alu (SVA) families specifically characterized the ALS-TE group. Remarkably, as the authors observed, ALS-TE subgroup was characterized by the lowest TARDBP expression. Additional pathways consistent with TDP-43 functions, such as the depletion of spliceosome and proteosome-linked genes were specifically depleted in the ALS-TE subgroup. Transcriptionally altered epigenetic regulators, namely, chromodomain-helicase-DNA-binding protein 5 (CHD5), lysine acetyltransferase 2A (KAT2A), and the histone H3K4 lysine methyltransferase 2B (KMT2B), were also part of this subgroup [138], in line with the impact of TDP-43 on global histone modifications reported above (see Section 2.1: TDP-43 Is a Global Epigenetic Modifier). The other subgroups displayed either more sustained alterations in the oxidative stress markers, including SOD1 mRNA (61%), or a strong bias for inflammation and pan-glial cells activation (19%). These conclusions were broadly supported by analyses performed in human SH-SY5Y neuroblastoma cells, where the use of CLAM, a tool designed to handle repetitive reads on sequencing data of TDP-43-bound RNA obtained by using enhanced cross-linking and immunoprecipitation (eCLIP-seq), unmasked 439 TE-derived RNA bound to TDP-43, corresponding to 31% of all mapping reads [138]. Specifically, 58% of the TE associated peaks (17.6% of the whole TDP-43 bound RNAs) mapped anti-sense with respect to the TEs, as already observed previously specifically for LINE-1 and Alu elements [35][139][35,139], and could provide regulatory sequences for the host genes they lie within [138]. In addition, knocking down TDP-43 using an shRNA in SH-SY5Y altered the expression level of several TE, mainly from the LTR class. All the significantly altered retrotransposon transcripts were upregulated, and only a fraction of which was identified by eCLIP-seq under normal TDP-43 expression [138], thus providing further evidences that TDP-43 normally contributes to the silencing of retrotransposon transcripts, and that this can be achieved at the RNA and DNA level. All these data support a specific functional and conserved role for TDP-43 in the repression/regulation of TE elements. Importantly, misregulated TE expression can have a number of detrimental impacts on chromatin, such as those observed in ALS and other neurodegenerative diseases. They may include genome instability via the spurious integrations of new repeats, activation of the DNA-damage stress response, or deregulation of the neighboring genes (Figure 3A).2.5.1. The Singular Case of Human HERV-K Env Protein Activated by TDP-43
An increased expression of the primate specific ERV-K family in a subgroup of sALS patients was reported in at least two studies [140][141][140,141], and TDP-43 was proposed to behave as an activator rather than a repressor. HERV-K overexpression of selected HERV copies was observed specifically to occur in the cortical and spinal neurons of some of the sALS patients, but not of healthy individuals, AD, or PD patient brains. The first study, looking for pol gene containing mRNA, identified several actively transcribed loci in the HERV-K HML-2 and 3 subfamilies, including specific copies with protein coding potential lying within a candidate interval for MND, in which the susceptibility genes were not identified [140]. Expression of the RT protein was observed specifically in ALS brains and localized to cortical and motor neurons [140]. In the second study, starting from an env perspective [141], an increase in ALS patients of env containing transcripts specifically from the family HERV-K was found. Immunostaining confirmed the high expression of HERV-K env protein in the cytoplasm of pyramidal cortical and spinal neurons in these ALS patients, but not in glial cells and not in healthy or AD affected tissues [141]. Further suggesting a possible connection with TDP-43, the ectopically driven TDP-43 expression in human stem cells-derived neurons increased the expression of in all gag, pol, and env retroviral genes regions in a correlated dose-dependent manner, supporting the activation of proviral forms of HERV-K [141], or the activation of multiple copies holding the same 5′LTR regulatory sequences. Mechanistically, the knockdown of endogenous TDP-43 with siRNA reduced HERV-K expression, thus arguing against a derepression of a HERV-K copy (or copies) caused by overexpression-mediated nuclear depletion of TDP-43 in the neurons of these patients or in vitro. In addition, ChIP assays, together with in vitro luciferase assays on the HERV-K LTR in HeLa cells, confirmed the activating effects of TDP-43 load and binding onto the LTR [141]. Furthermore, TDP-43 binding correlated with association of RNA Pol II p-Ser2, a processive form, on the consensus LTR from the HML-2 LTR5Hs-holding LTR-type subgroup. Interestingly, high affinity binding happens at a non-canonical polypyrimidine track (5′CCCTCTCCC) within the LTR region (+726), and less strongly at four other polypyrimidine motifs along it [141]. It is to be noted that the ChIP assays were performed on a plasmid holding a prototypal HERV-K LTR, and a final confirmation is required that the HERV-K elements under study are targeted by TDP-43 in their specific chromatin context within the genome. Expression of HERV-K, notably the env product, either by transfection of the prototypal HERV-K genome or HERV-K env gene into human neuronal cultures, or through transgenic mice expressing HERV-K env gene under the pyramidal neurons expressing Thy-1 promoter at a similar or higher level as observed in ALS patients, all caused neurotoxicity [141], triggering the degeneration of motor neurons and affecting the length, branching, and complexity of the dendrites as well as the number and the morphology of the spines. In the frontal cortex of the transgenic mice, yH2A.X foci were increased in neurons, and astrocytosis was noted in the surroundings, highlighting ongoing neuronal injury [141]. Interestingly, Cas9-directed downregulation of HERV-K env naturally produced in the prostate cancer cells LnCAP can trigger a strong diminution of TDP-43 mRNA and protein levels [142], suggesting the existence of a mutual activation loop between TDP-43 and HERV-K proviruses encoding env ORF. This system could be leveraged to downregulate TDP-43 overexpression. A schematic figure illustrating these findings is presented in Figure 3C. At this stage, however, and as underlined by Douville and colleagues [140], it remains unclear if the recombination of various HERV-K proteins originating from multiple loci may activate cycles of retrotransposition (or reinfection) and result in DNA damage leading to cell death. In addition, the youngest HML-2 family members, i.e., those belonging to the LTR5Hs that hold the activating TDP-43 binding site, present some degree of insertional and structural polymorphism in the population [117][118][119][117,118,119]. Furthermore, besides the potential retroviral ORFs production, LTR5Hs LTR elements have been shown to regulate hundreds of “cellular” genes [143]. These are all aspects of the TDP-43 regulation of the HERV-K family that deserve further investigations with respect to neurodegenerative diseases in which TDP-43 functions and levels are described to be altered.2.5.2. TDP-43 Control of LINE1 Retrotransposition
Heterochromatic regions are typically enriched for different types of intergenic repeats and depleted from genes. In agreement, the loss of heterochromatin identified in post-mortem human ALS brain nuclei without TDP-43 by Liu and colleagues via ATAC-seq was enriched for a particular class of interspersed intergenic repeat, i.e., the LINE1 elements [65]. Importantly LINE1 decondensation was not only accompanied by an increase in L1 transcription, indicative of their derepression, but also by increased LINE1 DNA in neuronal cells, meaning an increase in the number of LINE1 copies within the genome [65]. Functional LINE1 elements have the capacity to insert neo-retrotranscribed copies of themselves in ectopic places of the genome, a sort of “copy–paste” mechanisms. Hence, the authors of this study demonstrated that nuclear TDP-43-lacking cells displayed an increase in LINE1 retrotransposition [65]. In keeping with this view, in vitro experiments directly correlated the lack of TDP-43 in HeLa cells’ nuclei with a decrease in H3K9me3 histone heterochromatin modification and an increase in retrotransposition activity. This cell population was neuronal and corresponded to 7.05% of them on average and less than 2% of all cells [65]. These results suggest that there may be an accumulation of LINE1 nucleic acids in TDP-43-negative nuclei, which can potentially increase LINE1 DNA content, even in the absence of complete retrotransposition leading to truncated L1 [135]. Increases in L1 and/or in ERV RT are both consistent with the previously reported increase in RT activity in serum of HIV-1-negative ALS patients [144][145][144,145]. A new study, this time conducted in the mouse germline, shows that TDP-43 plays also an important role in inhibiting L1 retrotransposition in mouse embryonic stem cells (mESCs) and preimplantation embryos [146]. In this study, it was shown that TDP-43 interaction with L1 open reading frame 1 protein (L1 ORF1p) is necessary in order to mediate this genomic protection. It is important to note that this process is developmentally regulated; L1 retrotransposition is highly active in mammalian pre-implantation embryos [146]. Furthermore, an L1 retrotransposition assay in HEK293T cells revealed that deletion of the TDP-43 C-terminal domain severely compromised the inhibition of L1 retrotransposition, while RRM or NLS mutants retained their inhibitory capacity [146]. Therefore, in the brain of ALS patients, TDP-43 alterations may lead to LINE-1 reactivation via H3K9me3 reduction and chromatin decondensation, but in some instances, they could also be directly linked to the increase in L1 new copies integrated into the neurons genome. It is interesting to stress that in another pathological condition, i.e., a mouse model of progeria, L1 RNA was shown to negatively regulate the enzymatic activity of the H3K9me3 SUV39H1, thus sustaining heterochromatin loss [147]. A schematic illustration of these findings is presented in Figure 3E.2.5.3. Conservation of TDP-43/TDPH Regulation of TE in Drosophila
Flies have often represented a good model for TDP-43 deregulation and ALS. For this reason, TDP-43 impact on TE was recently examined ([148][149][150][148,149,150]. While the ERV-K family is not present in Drosophila, hTDP-43 overexpression in different brain cells of Drosophila (including neurons and glia) affected the expression of TE elements, principally the LTR and LINE classes, and generally triggered their activation [148][149][148,149]. The same was observed following TBPH loss in TBPH-null fly head tissues (TBPH being the homologue of TDP-43 in flies) [150]. Specifically in glial cells, this led to the activation and to the replication of ERV-related Gypsy retrotransposon, which appeared to be responsible for a substantial portion of the toxicity observed upon hTDP-43 overexpression [148][149][150][148,149,150]. Notably, both non-cell-autonomous propagation of DNA damage and apoptosis experienced by the adjacent neurons could be blocked by the Gypsy ERV glial silencing [148][149][148,149]. When investigating the siRNA pathways, which is a well-known mechanism of TE silencing, researchers noted that hTDP-43 expression interfered with siRNA-mediated—but not the miRNA-mediated—silencing, resulting in the desuppression of a reporter expression [148]. In glial cells, the reduction in siRNA silencing efficacy was marked and rapid, while in neurons, it was progressive and age-dependent [148]. Again, similar findings were obtained in TBPH-null Drosophila [150], and it was further found that TBPH interacts with the RISC component Dcr-2 mRNA and protein, regulating both its levels and activity [150]. This indicated an additional mechanism by which TDP-43 pathology could lead to TE silencing erosion and genome instability, including TE-mediated DNA damage. A schematic illustration of these findings is presented on Figure 3D. Although, siRNAs and miRNAs in Drosophila are processed largely via distinct pathways—Dcr2/Ago2 and Dcr-1/Ago, respectively—in mammals, the same DICER and Argonaute proteins process both miRNAs and siRNAs [151], a process in which TDP-43 has been shown to be implicated for at least a subset of miRNAs (see the review of [152]). In humans, suppression of TDP-43 in the neuroblastoma SH-SY5Y cells was found to produce a similar reduction in the human Dicer protein levels [150]. Furthermore, in the germ line, another type of siRNA using the ping-pong pathway (piwiRNAs and miwiRNAs) in mice and Drosophila derives from a large group of retrotransposons, which themselves and are linked to retrotransposon silencing and DNA methylation [153]. Similar mechanisms using the endo-siRNA pathways can drive LINE-1 DNA re-methylation in human breast cancer cells [154]. Interestingly, reverse-transcriptase inhibitors alone (stavudine, azidotimidine, tenofovir, or rilpivirine) has also been demonstrated to be effective in partially reverting the locomotion defects in TDPH-deficient flies induced by RTEs activation, with azidotimidine been the most efficient. Enotaxin, a compound capable of activating the siRNA pathway and able to counteract RTE activation, was also able to restore the locomotive behaviors and the formation of neuromuscular synapsis [150].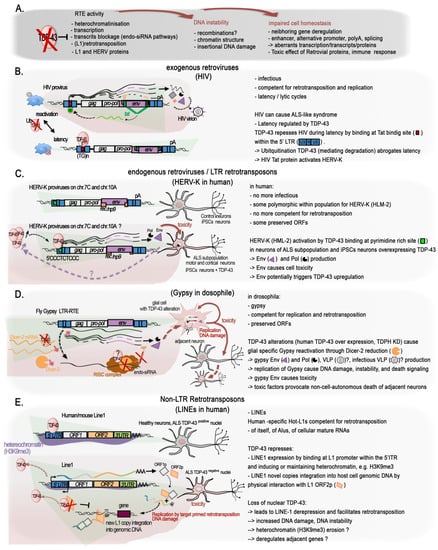
2.6. ALS and Epigenetic Functionality of TDP-43 Short Splicing Isoforms
Naturally occurring splice variant isoforms of TDP-43 leading to shorter TDP-43 proteins have been revealed on several occasions since the early 2000s [156][157][156,157], but their possible relevance in ALS has just started to be investigated. They all derive from the different use of close splice donors/acceptors sites within the exon 6 in the 3′ end of the TDP-43 ORF and part of the 3′UTR, creating a sixth intron (Figure 4A). The resulting proteins share at least AA1-AA256 with TDP-43, but they have an alternative C-terminus, lacking the highly disordered Glycine-rich region, and gaining an additional 18 AA sequence at the C-terminal end that is not present in wild-type TDP-43.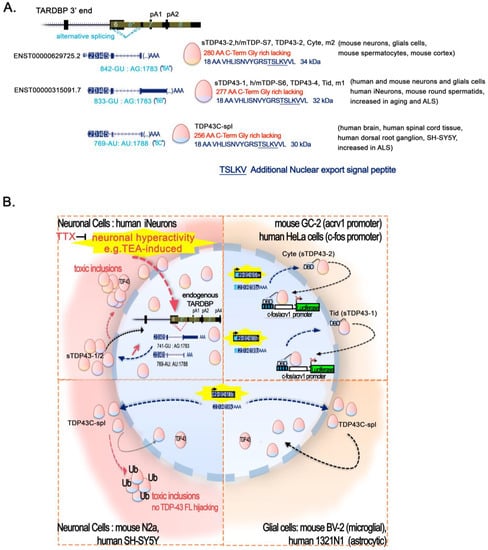
2.7. Epigenetic Role of TDP-43 Alternative Forms
Another intriguing finding of the work performed by Giannini and colleagues is the presence of the TDP-35 form at chromatin at R-loop, as highlighted by co-IP in both whole-cell lysates and chromatin fractions of lymphoblastoid cell lines (LCL). Regarding the importance of TDP-43 disease-associated mutations, it is interesting to note that this interaction was higher in for LCL carrying the A382T mutation of TDP-43 [97]. The authors initially understood TDP-35 to correspond to CTF35, a C-terminal fragment of 35 kDA, resulting from the truncation TDP-43 [97] using an antibody against AA203-209 [161]. However, the caspase induction of TDP-43 into cytotoxic CTF-35 is generally known to happen in the cytoplasm and to accumulate in detergent insoluble fraction. Furthermore, caspase-generated CTF-35 fragments have a disrupted nuclear localization signal (NLS), making them unlikely to travel back to the nucleus. In addition to these events, shorter-than-35-kDa TDP-43 immunoreactive products are numerous and can result from cleavage by other enzymes or potentially derive from alternate ORFs. In particular, analysis of Neuro2a cell lysates evidenced that Ca2+-activated calpain cleavage produces N-term fragments identified to be of about 36, 34, and 32 kDa [162]. Another work comparing caspase- and calpain-generated TDP-43 fragments via in vitro protease digestion of produced full-length TDP-43 shows a more complex pattern of cleavage product [163]. Notably, Caspase-3 induced 33 kDa fragments, whereas calpain produced 35 kDa fragments, and both could lead to the generation of 25 kDa fragments. Accordingly, calpain-I and caspase-3 cleavages have been shown to lead to several fragments being recognized exclusively by antibodies raised against either the N-term, the C-term, or internal epitopes [162][163][162,163]. This complexity highlights the difficulties in interpreting the biology of TDP-43 regulation and function, and the crucial importance of the tools used to identify TDP-43 fragments, as already reported. Notwithstanding these challenges, the results presented in both studies have highlighted the possibility that the chromatin associated fragment identified by Giannini and colleagues [97] could be an NTF of about 35 kDa rather than the thoroughly described CTF35. On top of this, another recent work has described TDP35 in the nucleus, detected using a C-term antibody (Gly 400 epitope). The authors of this work reported that this CTF35 was produced by the activity of caspase3 on TDP-43 in the nucleus, and that this proteolytic cleavage could be impeded by Malat-1 lncRNA binding to TDP-43 in the nucleus [164]. In any case, the fragment observed by Giannini and colleagues has been reported to exist in association with the RNA–DNA hybrid (detected by S9.6 antibody) on the chromatin, as well as in Neuro2a cells in the absence of calpain-I treatment, along with the 35 kDa TDP-43- related products [162]. Finally, in addition to all these species, it has been postulated that human and mouse cortices also show reactivity for N-term TDP-43 antibodies at 32.5 kDa and about 35 kDa of size, possibly corresponding to sTDP43-1 and sTDP43-2, fragments upregulated by neuronal hyperactivity [158][159][158,159] or to the recently identified sTDP43C-spl [160]. Indeed, all these alternatively spliced isoforms, as mentioned in the previous (Section 2.6: ALS and Epigenetic Functionality of TDP-43 Short Splicing Isoforms) section, have been shown to localize in the cytoplasm or in the nuclear space either on chromatin or in speckles, depending on the various experimental conditions. Regardless of the effective(s) scenario, the presence of short TPD-43 products—either splicing isoforms or proteolytic cleavage-products of full-length TDP-43—in the nucleus and, in particular, on the chromatin, will certainly require further investigation pertaining to their functional output in physiological and pathological situations.2.8. Epigenetic Landscape Modifications Associated with TDP-43 Mutants
TARDBP mutations have been identified mostly in familial ALS patients but also in sporadic FTD, AD, and PD cases, and more than 50 TDP-43 variants have been linked to the incidence of ALS/FTD [165][166][165,166]. They are mostly found in the C-term part of the protein, while in the N-term, A90V in the NLS and P112H and D169G have been reported in the RRM1 motif [165]. Functional observations have been made for about twenty of ALS-mutants, as reviewed in [165][166][165,166] and for mouse and in vitro models holding TDP-43 mutations, as described in [167][168][169][167,168,169], reporting altered normal RNA splicing with or without concomitant cytoplasmic aggregation. Few TDP-43 models carrying null or point mutations have been investigated to explore the association of pathological TDP with altered epigenetics treats. Depending on the regions hinted by mutations, different scenarios on chromatin impact can be envisaged. Indeed, mutations in the C-term—known to guide nuclear loss—are expected to have a broad impact on TDP-43 functions both at the RNA and the DNA levels. This might naturally hold true for mutations within the NLS or the NES controlling the nucleocytoplasmic shuttling. Instead, mutations in the N-term, whether at the RRM1 or RRM2, may have a more focalized impact on the RNA-processing aspect or on chromatin, whether linked to RNA processing aspects or lncRNA-related functions of TDP-43 on chromatin, but also on TDP-43′s ability to bind ssDNA or dsDNA, as has been documented in different studies [15][17][19][20][21][22][15,17,19,20,21,22], and notably regarding free dsDNA ends [23]. Such mutations might impact either transcriptional regulation, the ability to bind DNA damaged ends, or the ability to restrict TE activity. For example, mutations of the TDP-43 RRM1 that abolish its DNA/RNA binding, as shown with F147/149L, can be expected to modify TDP-43-mediated gene expression. Consistently, as reported above, mutants either lacking RRM1 motif or with a mutated form (F147L/F149L) are sufficient to disrupt TDP-43 repressive function on the Acrv1 promoter [40] and on its own promoter [170]. In addition, RRM1–RRM2 acetylation-mimic point mutations (KK-QQ), but not acetylation-null (KK-AA), abolished CHOP transcriptional activation [48]. Instead, deletion of the TDP-43 C-terminal domain severely compromised the inhibition of L1 retrotransposition, while RRM or NLS mutants maintained their inhibitory capacity [141]. Expression of hTDP-43 carrying a mutated nuclear localization signal (ΔNLS-hTDP-43; [63][64][63,64]) conveyed notable changes in gene expression, including a dysregulation of histone 3′ end-processing machinery paralleled by an increased canonical histone transcript, reinforcing the remarkable role that TDP-43 has in the function of the chromatin assembly pathway. All ALS/FTD TDP-43 mutations reported to date to have an impact on chromatin are concentrated in the C-term region and lead to either a defect in DNA methylation or DNA damage or to both, and some also impact TDP-43 own regulation (Table 2).| TDP-43 Mutation |
Epigenomic Alterations | Cellular Context | Refs. | |
|---|---|---|---|---|
| G294V | DNA damage/DSB TC-DSB | Mislocalization; | SH-S5Y5 | [97] |
| R-loop accumulation; R-loop-dependent increased DSBs | ||||
| Accumulation of FANCD2 repair foci (replication blockage). | ||||
| G298S | DNA methylation |
Hyper- and hypo-methylated regions related to controls with common and specific DMRs related to other ALS hIPSCs-derived MNs (C9orf72-, TARDBP-, SOD1-, and FUS-mutation carriers). | hIPSCs-derived MNs from G298S carrier | [74] |
| A315T | Chromatin remodeling | Diminution of nBAF chromatin-remodeling complex components (Brg1, BAF53b, and CREST). | Cultured mouse MNs | [57] |
| DNA methylation |
Altered 5mC and 5hmC underlying their increased vulnerability to degeneration. | TDP-43A315T mouse CSMNs | [171] | |
| DNA damage/DSB | Enhanced vulnerability to DNA damages. | MN-like NSC-34 cells | [95] | |
| M323K | Autoregulation | Increased RNA, but not significant for protein. | Mouse TDP-43M323K knock-in | [167] |
| Q331K | Autoregulation | Increase in RNA and nuclear protein in frontal cortex and spinal cords of mutant mice, but not in motor neuron. | Mouse TDP-43Q331K knock-in but not transgene | [168][169][168,169] |
| DNA damage/DSB | Increased cytosolic sequestration of the poly-ubiquitinated and aggregated form, nuclear loss of function DNA damage induction, and DSB repair defects (preventing the nuclear translocation of XRCC4); Contribution to oxidative genome damage accumulation via increased reactive oxygen species (ROS). |
Spinal cord of ALS Q331K carriers, SH-S5Y5 |
[93] | |
| Enhanced vulnerability to DNA damages (increase yH2Ax post damaging agent exposure). |
MN-like NSC-34 cells | [95] | ||
| M337V | DNA methylation |
No global alteration in 5mC | SH-S5Y5 | [58] |
| Histone PTMs | Significant decrease in global H3S10Ph-K14Ac; No significant increase in H3K9me3. |
|||
| DNA damage/DSB | Impairment in the NHEJ DSB repair factors recruitment. | Fibroblasts of human M337V carrier | [95] | |
| G348C | Autoregulation | Upstream intron1 promoter region of TARDBP binding but less efficient than WT for its repression; No significant activation of the intron 1 promoter. |
HEK293T | [170] |
| A382T | DNA Methylation |
No global alteration in 5mC. | SH-S5Y5 | [58] |
| Histone PTMs | No significant decrease in global H3S10Ph-K14Ac; No significant increase in H3K9me3. |
|||
| DNA damage/DSB TC-DSB | Increased yH2Ax; R-loop accumulation; R-loop-dependent increased DSBs accumulation of FANCD2 repair foci (replication blockage). |
SH-S5Y5 LCL from A382T carriers |
[97] | |
| Isoforms/fragments | Increased TDP-35 form at chromatin at R-loop. | LCL from A382T carriers | [97] | |
| Autoregulation | Upstream intron1 promoter region of TARDBP binding but less efficient than WT for its repression; Able to induce a significant activation of the intron 1 promoter. |
HEK293T | [170] | |
| N390D | DNA methylation |
Hyper- and hypo-methylated regions related to controls with common and specific DMRs related to other ALS hIPSCs-derived MNs (C9orf72-, TARDBP-, SOD1-, and FUS-mutation carriers). | hIPSCs-derived MNs from N390D carrier | [74] |
| G418C | Chromatin remodeling | Diminution in nBAF chromatin-remodeling complex components (Brg1, BAF53b and CREST). | Cultured mouse MNs | [57] |
| C9orf72 with loss of nuclear TDP-43 | DNA methylation |
Altered DNA 5mC and 5hmC in the CSMN without nuclear TDP-43 compared to residual C9orf72 CMSN without TDP-43 nuclear loss. | Residual lower MNs from post-mortem CSMN tissues of C9orf72 carriers | [73] |
