Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Fanny Huang and Version 1 by Nico M. Van Straalen.
Nonribosomal peptide synthetases (NRPSs) synthesize a range of peptide products with a wide spectrum of biological functions including antibiotic and siderophore activities. They are used in industrial biotechnology to produce various pharmaceuticals such as cytostatics and immunosuppressants. NRPSs are widespread among both prokaryotes and eukaryotes.
- NRPS
- nonribosomal peptide synthetases
- animals
1. The Distribution of NRPS Genes in Animals
Genomic data were available for 19 animal phyla, with most genomes representing Chordata, Nematoda, and Arthropoda. Researchers identified NRPS clusters in over 8% of the 1059 animal genomes analyzed. For each NRPS cluster identified by antiSMASH, researchers conducted a contamination check to analyze whether the putative NRPSs were truly part of the animal genome by evaluating whether the surrounding genes had best BLAST hits to other animals. AntiSMASH identified 772 putative NRPS clusters, of which 199 passed the contamination check and are likely true animal sequences. The occurrence of NRPSs showed a scattered distribution over metazoan phyla: Annelida, Arthropoda, Chordata, Cnidaria, Echinodermata, Hemichordata, Mollusca, Nematoda, Nemertea, Phoronida, Platyhelminthes, Priapulida, and Rotifera. Within the phylum Nematoda, genomes were available from Rhabditina, Tylenchina, Spirurina, Plectida, and Dorylaimia. NRPS and NRPS–polyketide synthases (NRPS–PKS) were identified in many species, but not in the early-diverging Dorylaimia.
Figure 1 shows the phylogenetic relationships among all animal NRPS sequences. NRPSs from Platyhelminthes clustered with Ebony as expected. The A domains from Annelida, Priapulida, Hemichordata, Nemertea and Cnidaria identified using Sm-NRPS were also part of this clade (Figure 1). In this clade, researchers also found Branchiostoma belcheri and Branchiostoma floridae (Chordata). In contrast to Ebony proteins, the Branchiostoma proteins did appear to contain regular condensation domains. A rotifer NRPS that showed similarity to Sm-NRPS contained one adenylation domain that clustered with Ebony and two adenylation domains that clustered with NRPSs identified by antiSMASH.
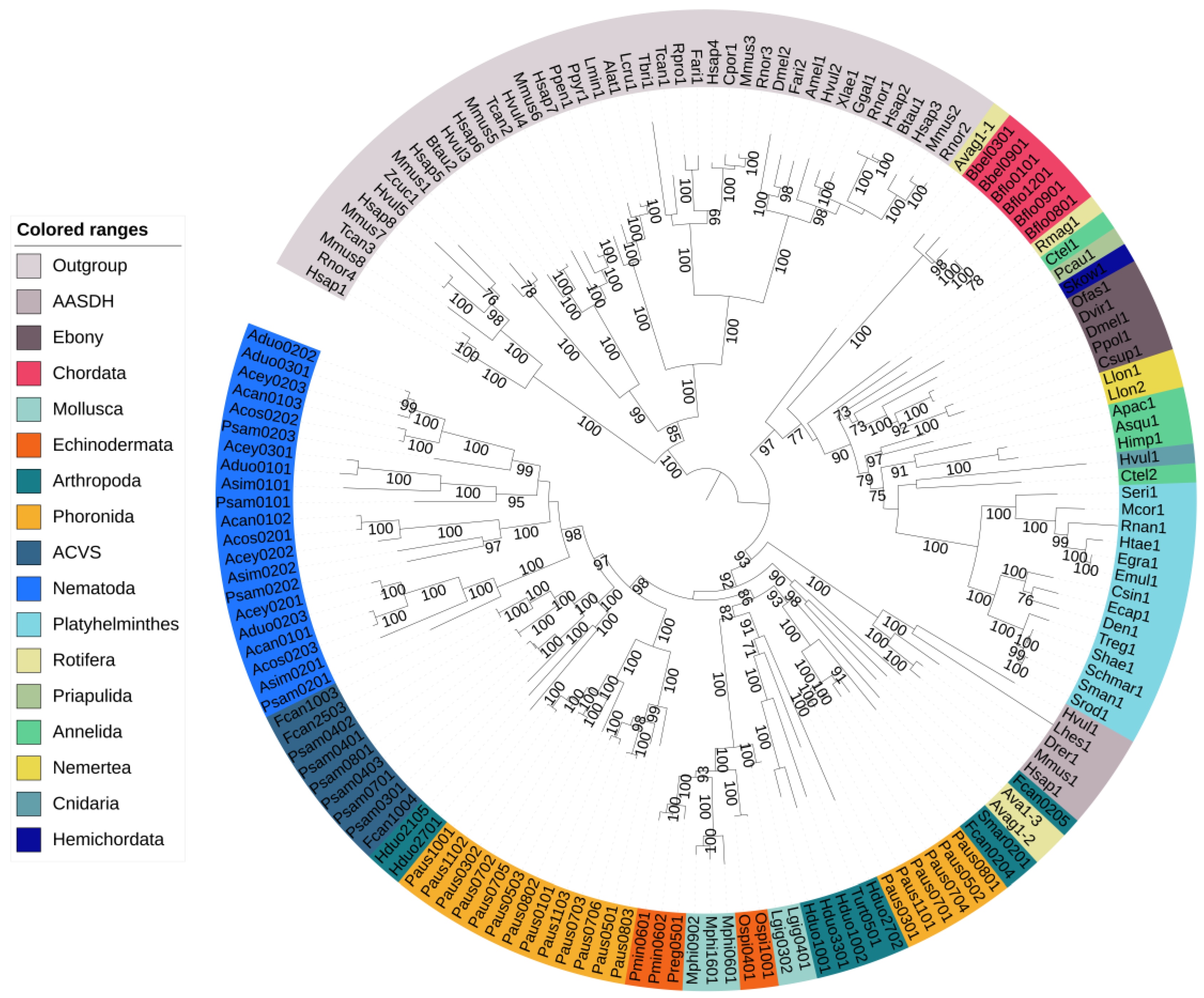
Figure 1. Phylogenetic tree of putative nonribosomal peptide synthetases and other adenylate-forming enzymes in animals. The outgroup proteins formed a monophyletic clade. Ebony clustered with NRPSs from Platyhelminthes and other proteins identified based on their similarity to Sm-NRPS. The chordata are also part of this clade. AASDH clustered with putative NRPSs identified by antiSMASH. NRPSs identified in Mollusca and Echinodermata were found in a single clade in the tree. The nematode NRPSs formed a monophyletic clade except for several Plectus sambesii adenylation domains, which clustered in the ACVS clade. Bootstrap support ≥70% is shown.
The NRPSs in Nematoda formed a monophyletic clade except for some Plectus sambesii adenylation domains. This is discussed below under beta-lactam biosynthesis genes in P. sambesii. In the springtail F. candida (Arthropoda), researchers identified a second NRPS, in addition to the two copies of ACVS reported previously, on scaffold LNIX01000001.1. Furthermore, in another springtail, Holacanthella duospinosa, researchers also found an NRPS that was different from ACVS.
The NRPSs identified in Echinodermata (Ophiothrix spiculata, Patiria miniate, Patiriella regularis) and Mollusca (Lottia gigantea, Modiolus philippinarum) formed a monophyletic clade, even though the phyla themselves were not related at all (echinoderms are deuterostomes, molluscs are protostomes). It should be noted that all NRPSs in molluscs and echinoderms are located on short scaffolds, which makes it more difficult to achieve absolute certainty regarding the absence of contamination. Lastly, a clade of arthropod NRPSs consisting of Tetranychus urticae (Acari) and Holacanthella duospinosa (Collembola) was present in the tree.
2. Putative NRPSs Are Abundant in Rotifers
In rotifers, researchers identified up to 79 NRPS clusters per species (Table 1). One of these clustered with non-NRPS adenylating enzymes in the phylogenetic analysis (Figure 2). The putative NRPSs of the four rotifer species did not cluster in separate groups, but instead formed several clades together. researchers found both pure NRPSs and NRPS–PKS hybrids in rotifers. Moreover, large differences were observed between species in both the number of NRPS clusters and their predicted substrates (Table 1 and Figure 3). No NRPSs were identified in Brachionus calyciflorus, a rotifer that is classified outside the subclass Bdelloidea, to which the other four rotifers belong.
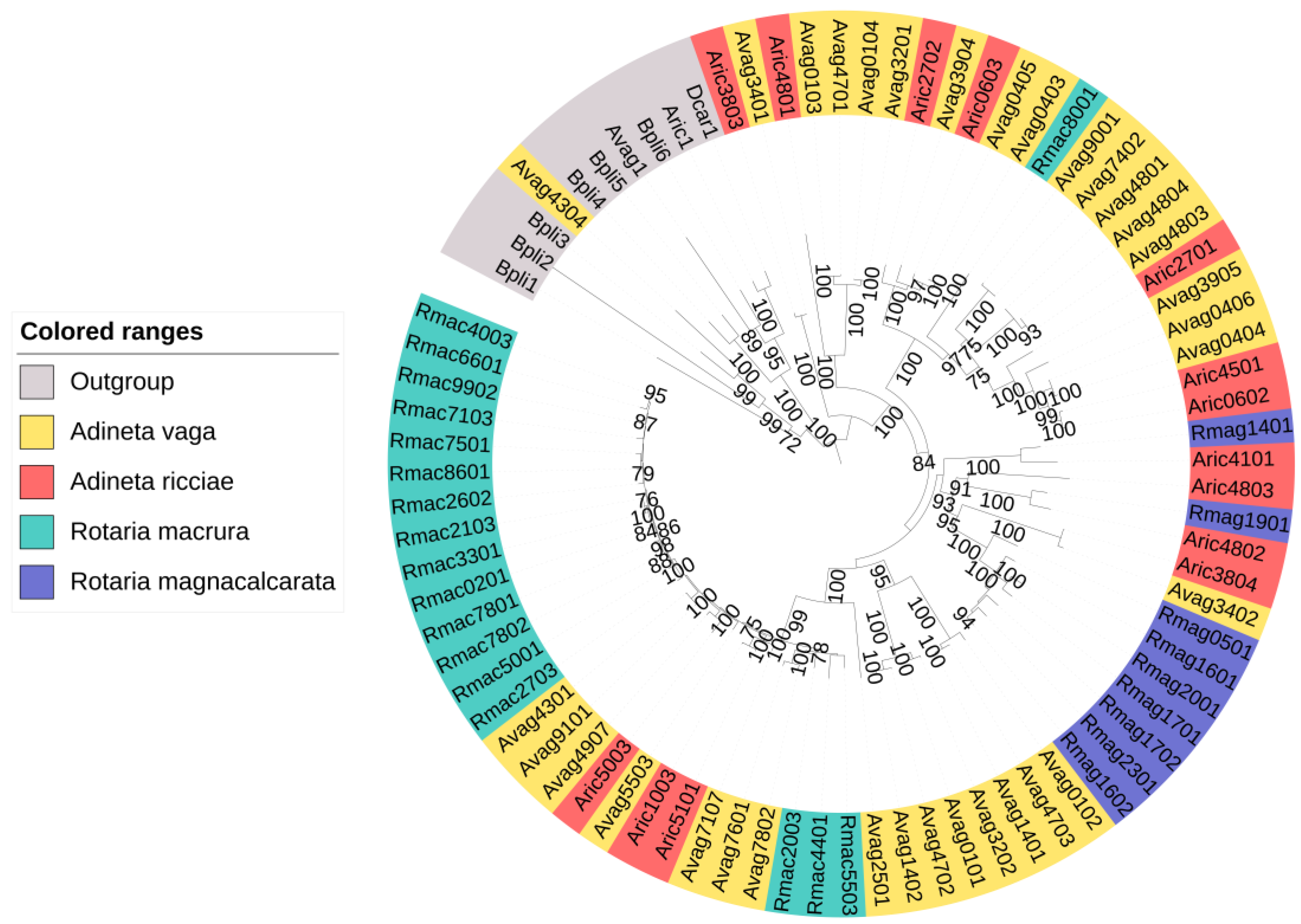
Figure 2. Phylogeny of the identified putative rotifer NRPSs and other rotifer adenylate-forming enzymes (outgroup). One of the putative NRPSs clustered with the outgroup proteins. The others formed separate clades. Bootstrap support ≥70% is shown.
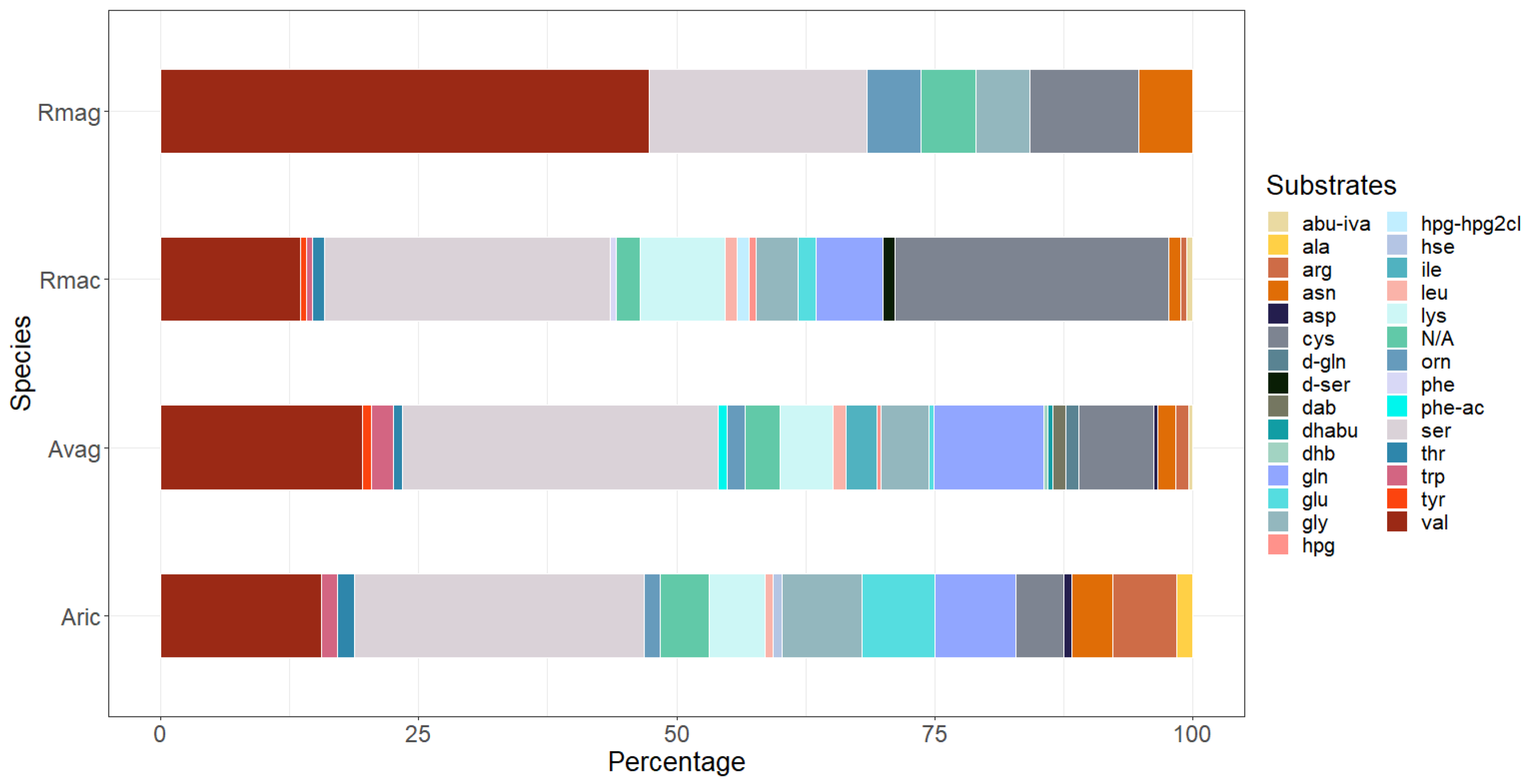
Figure 3. Substrate predictions for putative nonribosomal peptide synthetases (NRPSs) in rotifers. Rotifer NRPSs are predicted to use a wide variety of amino acid substrates including nonproteinogenic amino acids. Abbreviations: Rmag: Rotaria magnacalcarata; Rmac: Rotaria macrura; Avag: Adineta vaga; Aric: Adineta ricciae.
Table 1. Nonribosomal peptide synthetases (NRPSs) and hybrid nonribosomal peptide synthetase–polyketide synthases (NRPS–PKSs) in rotifers. The bdelloid rotifers from the genera Adineta and Rotaria contained many NRPSs, while they appeared to be completely absent in the genome of the non-bdelloid Brachionus calyciflorus.
| Species | NRPS | NRPS–PKS |
|---|---|---|
| Adineta ricciae | 46 | 2 |
| Adineta vaga | 69 | 5 |
| Rotaria macrura | 79 | 0 |
| Rotaria magnacalcarata | 12 | 1 |
| Brachionus calyciflorus | 0 | 0 |
It should be noted that some rotifer NRPS clusters were rather short and might have in fact together formed a single NRPS. This would reduce the total number of NRPSs reported in Table 1. The smaller NRPS clusters are often located on small scaffolds that do not contain other genes. However, for Adineta vaga, a new chromosome-level assembly has been published [26][1], which contains most of the NRPS clusters that were identified here in the more fragmentary assemblies.
3. Beta-Lactam Biosynthesis Genes in Plectus sambesi
Several of the adenylation domains of the nematode P. sambesii clustered with springtail L-δ-(α-aminoadipoyl)-L-cysteinyl-D-valine synthetase (ACVS) (Figure 1). ACVS catalyzes the first step of beta-lactam antibiotic biosynthesis in microorganisms. Hence, researchers analyzed whether the genome of P. sambesii also contained other enzymes from the beta-lactam biosynthesis pathways. In addition to ACVS, researchers identified an isopenicillin N synthase (IPNS), which catalyzes the second step of the pathway that forms the beta-lactam ring and a deacetoxycephalosporin C synthase (cefEF, DAOC synhase).
P. sambesii ACVS shares approximately 50% amino acid identity with F. candida ACVS, while P. sambesii IPNS has an identity percentage of approximately 65% to F. candida IPNS. The predicted substrates for the three adenylation domains of the P. sambesii protein are aminoadipic acid, cysteine, and valine, which are the expected substrates for all known ACVS proteins (Figure 4).
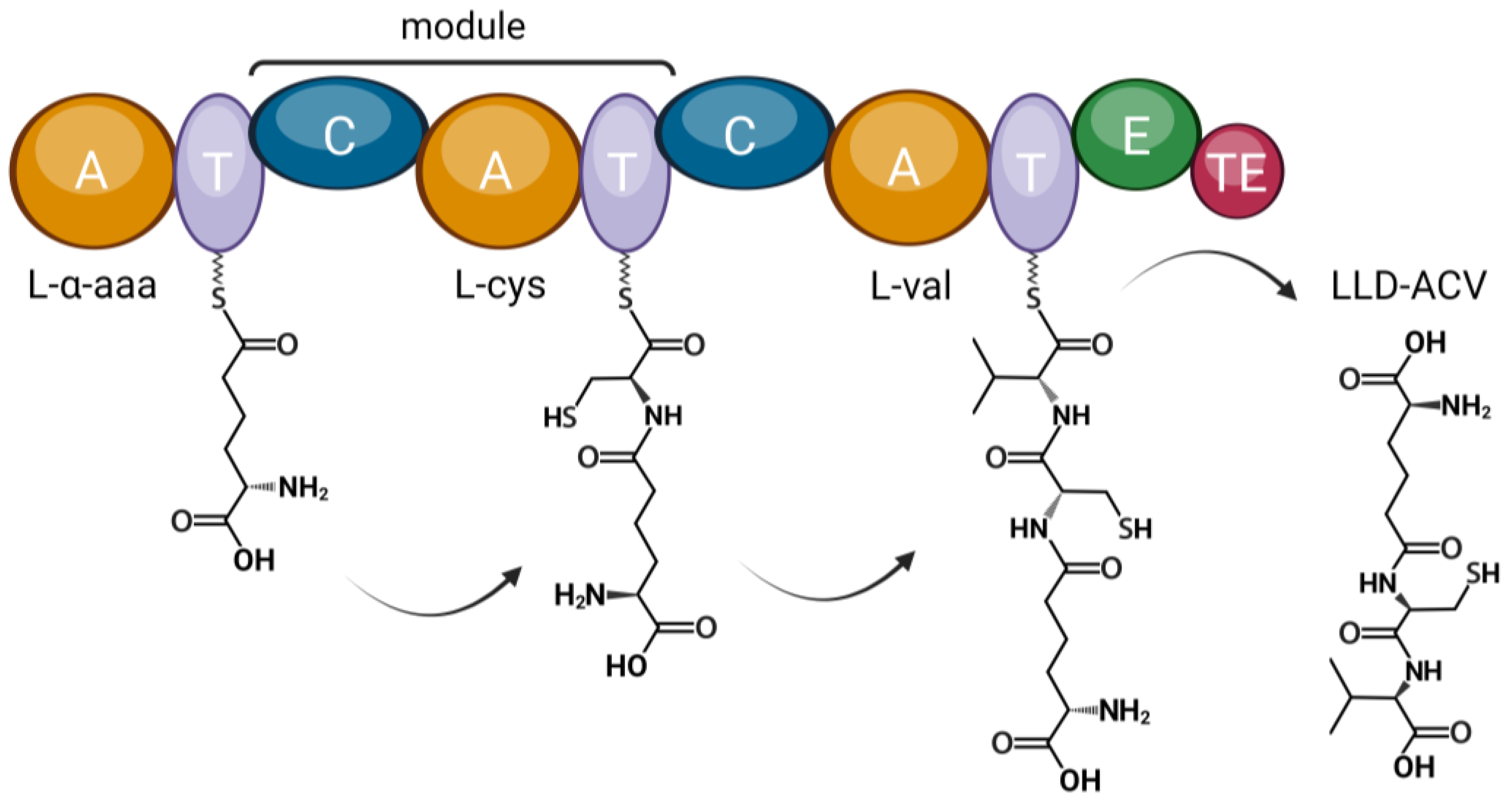
Figure 4. The domain architecture of ACVS. The structure and substrate predictions of a Plectus sambesii putative NRPS resemble that of bacterial, fungal and springtail ACVS. The predicted substrates based on the adenylation domains are: aminoadipic acid (aad), cysteine (cys), and valine (val). The final product is a tripeptide (LLD-ACV). The amino acid substrates are activated by the adenylation (A) domains and attached to phosphopantetheine groups that are bound to the thiolation (T) domains. Amide bonds between neighbouring substrates are formed by the condensation (C) domains. The epimerization (E) domain epimerizes L-valine while the thioesterase (TE) domain catalyzes the release of the peptide. Created with BioRender.com.
Many more animal genomes than previously assumed contain nonribosomal peptide synthetase genes. The capacity of animals to synthesize secondary compounds playing a role in signaling, antibiotic synthesis, and other functions might be much greater than thought before. This could also lead to discovery of new NRPSs with biotechnological applications. They are scattered across the metazoan tree and some clades contain many more NRPSs than others. These genes are often assumed to have entered animal genomes by horizontal gene transfer from bacteria, but since they are not restricted to specific lineages, it seems unlikely that all of them represent HGT events, although some do for sure. In addition, it is unclear which ecological traits predispose an animal to recruit these genes. A more detailed analysis, concentrated on specific lineages, could reveal the underlying mechanisms.
References
- Simion, P.; Narayan, J.; Houtain, A.; Derzelle, A.; Baudry, L.; Nicolas, E.; Arora, R.; Cariou, M.; Cruaud, C.; Gaudray, F.R.; et al. Chromosome-level genome assembly reveals homologous chromosomes and recombination in asexual rotifer Adineta vaga. Sci. Adv. 2021, 7, eabg4216.
More
