You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Catherine Yang and Version 1 by Irina Valerievna Pronina.
LncRNAs can alter gene expression and/or its functions by acting as miRNA spongers, via a direct interaction of lncRNAs with mRNAs or binding to RNA-binding proteins (RBPs). RBPs were shown to regulate mRNA expression and stability at the post-transcriptional level. RBPs combine a flexible structure with a versatile RNA-binding domain. These properties allow RBPs to engage in highly dynamic interactions both with other proteins as well as with coding and non-coding RNAs, leading to ribonucleoprotein complexes (RNPs) being formed. RNPs regulate RNA splicing, polyadenylation, stability, localization, translation, and degradation. In non-small-cell lung cancer (NSCLC), lncRNAs can regulate the levels and stability of target mRNAs by binding RBPs to form RNP complexes, as was demonstrated in a number of examples.
- lncRNA
- mRNA
- miRNA
- RNA-binding protein
- lung cancer
1. IGF2BP1/2/3 as RBP Mediator
IGF2BP1/2/3 (insulin-like growth factor 2 mRNA-binding protein 1/2/3) proteins are the most common RBPs mediating lncRNA regulatory functions. RBPs promote cancer cell proliferation, migration, and invasion, and their oncogenic functions are mediated by post-transcriptional regulation of mRNA stability and translation [1].
LINC01232 (long intergenic non-protein coding RNA 1232) is highly expressed in NSCLC cells and promotes cell stemness. This was suggested by estimating the mRNA and protein levels of stem cell markers (OCT4, Nanog, CD133, SOX2, and SOX4) and the sphere formation assay [2]. Two binding sites for the transcription factor FOXP3 (forkhead box P3) in the LINC01232 promoter region were predicted using DNA motifs from the JASPAR database. Luciferase reporter and ChIP assays were employed to confirm the direct binding between FOXP3 and the LINC01232 promoter, which activates LINC01232 transcription. LINC01232 was shown to activate the TGF-β signaling cascade by assessing the levels of the major proteins involved in this pathway (TGF-β1, α-SMA, SMAD2, and SMAD3) by Western blotting. StarBase (http://starbase.sysu.edu.cn/index.php, accessed on 19 June 2023) and the GEPIA database identified RBPs for LINC01232 and transforming growth factor beta receptor 1 (TGFBR1) as the critical modulators of the TGF-β signaling pathway, which could be activated by LINC01232. Three candidate RBPs (IGF2BP2, IGF2BP3, and DKC1) activated in NSCLC tissues and potentially associated with TGFBR1 and LINC01232 were selected for further analysis. Using RIP and RNA pull-down assays, IGF2BP2 was identified as the only RBP that interacted with both TGFBR1 and LINC01232. IGF2BP2 and LINC01232 were co-localized in the cytoplasm, as shown by FISH and FISH-IF assays. RT-qPCR and Western blotting revealed a positive association between LINC01232 and TGFBR1, indicating that IGF2BP2 overexpression can upregulate TGFBR1 expression, increasing both TGFBR1 mRNA and protein levels. In summary, FOXP3-activated LINC01232 in the RNP complex with IGF2BP2 was shown to increase the stability of TGFBR1 mRNA and therefore stimulate the TGF-β signaling pathway, facilitate the stemness of NSCLC cells, and induce the M2 polarization of macrophages [2].
The lncRNA MNX1-AS1, also known as LOC645249 and CCAT5 (colon-cancer-associated transcript 5), is an antisense transcript of the motor neuron and pancreas homeobox protein 1 MNX1. MNX1-AS1 is highly expressed in NSCLC and exhibits oncogenic properties [3]. Its transcriptional activation involves transcription factor c-Myc and the copy number amplification mechanism. MNX1-AS1 overexpression is associated with poor clinical outcomes in NSCLC patients. It is shown to promote cell proliferation and colony formation in vitro and tumor growth in vivo. In NSCLC, MNX1-AS1 binds to IGF2BP1 and causes phase separation of IGF2BP1, which facilitates the IGF2BP1 binding to the 3’UTR of c-Myc and E2F1 mRNAs and enhances the stability of these mRNAs [3]. These interactions form a positive feedback loop that drives IGF2BP1 phase separation and promotes c-Myc and E2F1 signaling, cell cycle progression, and proliferation of NSCLC cells (Table 1).
Table 1.
Action of lncRNAs mediated by RNA-binding proteins (RBPs) in NSCLC progression.
| Mechanisms, Axes | LncRNAs/Axes in Processes, Pathways, Prognosis, Survival, and Drug Resistance |
Ref. |
|---|---|---|
| IGF2BP1/2/3 as an RNA-binding protein (RBP) | ||
| LCAT1↑→m6A-IGF2BP2(RBP)↑stab →m6A-CDC6↑mRNAstab |
Promotes NSCLC cell growth, migration, and poor patient survival | [4] |
| Linc-SPRY3-2/3/4↓ov-ex+IGF2BP3(RBP) /HMGA2, c-MYC↓mRNAstab |
Suppresses NSCLC and enhances cell radiation response | [5] |
| FOXP3→(pr)LINC01232↑+IGF2BP2(RBP)→TGFBR1↑stab | Promotes TGF-β signaling, NSCLC cell stemness | [2] |
| IGF2BP2(RBP)↑→m6A-MALAT1↑stab→ATG12↑protein | Promotes NSCLC proliferation; reduces OS, DFS | [6] |
| c-Myc→MNX1-AS1↑+IGF2BP1(RBP) ↔c-Myc, E2F1↑mRNAstab→c-Myc-sign |
Promotes cell cycle progression, proliferation in vitro, in vivo; poor clinical outcomes | [3] |
| lnc-THOR↑→IGF2BP1(RBP)↑ →IGF2, Gli1, Myc, SOX9↑mRNAstab |
Enhances NSCLC cell proliferation, migration, and invasion | [7] |
| HuR/ELAVL1 as an RNA-binding protein (RBP) | ||
| FENDRR↓ov-ex/MDR1↓mRNAstab ↔MDR1↑3’UTR← HuR(RBP) |
Suppresses NSCLC cell stemness | [8] |
| E2F1→MCF2L-AS1↑+HuR(RBP)→CCND1↑mRNAstab | Drives NSCLC cell growth and induces gefitinib resistance | [9] |
| SNHG12↑+HuR(RBP)→USP8↑mRNAstab,protein →PD-L1↑mRNAstab, protein↑/CD8+T-cell↓ |
Promotes proliferation, migration, invasion, and immune escape in vitro/in vivo | [10] |
| Heterogeneous nuclear ribonucleoproteins (hnRNPs) with RBP function | ||
| SChLAP1↑/hnRNPD(AUF1, RBP)↓/PDL1↑mRNAstab | Enhances proliferation, immune evasion | [11] |
| DNA-meth/DIO3OS↓ov-ex/hnRNPK↓ /MYC,DNA,mRNA↓/CDC25A↓ |
Ectopic expression of DIO3OS suppresses NSCLC tumorigenesis, metastasis in vivo | [12] |
| LIMD1-AS1↓ov-ex+hnRNPU→LIMD1↑mRNAstab | Suppresses NSCLC progression | [13] |
| Other RNA-binding proteins (RBP) as mediators of lncRNAs | ||
| FAM83A-AS1↑+FBL(RBP)→FAM83A↑pre-mRNAstab | Promotes LUAC metastasis, ERK signaling; low OS, PFS | [14] |
| LINC00667↑+EIF4A3(RBP)→VEGFA↑mRNAstab | Promotes proliferation, migration, and angiogenesis | [15] |
| MACC1-AS1+UPF1(RBP)→LATS1/2↓mRNAdestab | Drives NSCLC cell stemness through inhibition of the Hippo pathway | [16] |
| TM4SF19-AS1↑+WDR5(RBP)→TM4SF19(pr-WDR5) →(DNA-demeth-pr) TM4SF19↑mRNA |
Facilitates proliferation, adhesion of lung squamous cell carcinoma | [17] |
| METTL3→m6A-DLGAP1-AS2↑stab +YTHDF1(m6A-reader RBP)→c-Myc↑mRNAstab |
Promotes aerobic glycolysis; correlated with advanced stages, poor prognosis | [18] |
Note: stab—stability; ov-ex—overexpression; demeth-pr—demethylated promoter; meth—methylation; DFS—disease-free survival; OS—overall survival; PFS—progression-free survival; p-w—pathway; phosph—phosphorylation; sign—signaling; ↑ ↓—increase or decrease in expression or stabilization; →—activation; /—inhibition; ↔—a positive feedback.
Ectopic overexpression of lnc-THOR in NSCLC elevated the expression of IGF2BP1 target mRNAs (IGF2, Gli1, Myc, and SOX9), enhancing its mRNA stability (Table 1). The direct binding of lnc-THOR to the IGF2BP1 protein was validated using RIP and RNA pull-down assays [7]. Through this axis, lnc-THOR stimulates the proliferation, migration, and invasion of NSCLC cells in vitro and tumor growth in vivo [7].
There is evidence for the specific contribution of linc-SPRY3 (sprouty protein homolog 3)-2/3/4, the Y chromosome transcript, to the radiation response of male NSCLC [5]. Predicting RBP motifs using a bioinformatic approach allowed for identifying IGF2BP3 as a binding partner for linc-SPRY3-2/3/4. Direct interaction of IGF2BP3 with linc-SPRY3-2/3/4 was validated using UV cross-linking and immunoprecipitation (CLIP) assays. Linc-SPRY3-2/3/4 reduced the stability of the mRNA of anti-apoptotic and oncogenic HMGA2 and c-MYC targets by binding IGF2BP3 and increased the radiosensitivity of male NSCLC [5]. Thus, suppressor lncRNAs linc-SPRY3-2/3/4 mediated with IGF2BP3 increased the response to radiotherapy on the Y chromosome (Table 1).
RBPs can also act as a N(6)-methyladenosine (m6A) reader or m6A writer proteins in mRNA methylation via (m6A)-methyltransferase complex [19][20]. LncRNA LCAT1 (lung-cancer-associated transcript 1) was shown to bind and stabilize the m6A-IGF2BP2 reader protein, preventing its lysosomal degradation [4]. Stabilized m6A-IGF2BP2 then facilitated the translation and stability of cell division cycle 6 protein (CDC6) mRNA, which promoted the proliferation, survival, and migration of NSCLC cells. RNA pull-down, mass spectrometry, and RIP were used to validate the direct interactions.
Using the CRISPR/Cas9 system, METTL3, the core m6A methyltransferase, was found to be involved in interactions between IGF2BP2 protein and CDC6 mRNA dependent on m6A modifications. Therefore, METTL3 (m6A-writer) and m6A-IGF2BP2 (m6A-reader) promoted the upregulation and stabilization of CDC6 by LCAT1, which enhanced the progression of NSCLC and impaired patient survival [18] (Table 1).
Another study showed an activating and stabilizing effect of IGF2BP2 as both RBP and m6A-reader in the axis with MALAT1 lncRNA and ATG12 (autophagy-related 12) protein [6] (Table 1). First, IGF2BP2 mRNA level was elevated in primary NSCLC tissues and positively correlated with poor overall survival (OS) and disease-free survival (DFS). Its ectopic expression and knockdown in NSCLC cell lines and in vivo showed that IGF2BP2 promoted NSCLC cell proliferation and tumor growth. We would like to emphasize that the activating and stabilizing effect of IGF2BP2 is exerted directly on the lncRNA MALAT1 as well as indirectly via the formation of m6A-MALAT1 by m6A-RNA methylation. Direct binding between IGF2BP2 and MALAT1 was validated using the RIP-PCR assay, while mRIP-PCR with m6A antibody confirmed the m6A modification of MALAT1. Western blotting showed that the overexpression of IGF2BP2 binding MALAT1 increased the level of ATG12 protein, a key downstream target of MALAT1 [6]. Thus, IGF2BP2 was demonstrated to enhance MALAT1 stability through m6A modification promoting the protein expression of its downstream target ATG12, thereby facilitating NSCLC progression and reducing patient survival (Table 1).
2. HuR/ELAVL1 as RBP-Mediator
Post-transcriptional gene regulation relies on hundreds of RNA-binding proteins. One of the mechanisms of post-transcriptional gene regulation in mammalian cells is the rapid degradation of mRNAs containing AU-rich elements (ARE) in their 3’-UTR [21]. ELAVL1 (Drosophila ELAV-like RNA-binding protein 1) or HuR (human antigen R) belongs to the ELAVL family of RBPs that contains several RNA-binding domains. Overexpressed HuR/ELAVL1 selectively binds to cis-acting ARE and stabilizes ARE-containing mRNAs in cells. Human RBP HuR/ELAVL1 is a conserved mRNA stability regulator [21][22].
HuR/ELAVL1 was reported as the common RBP of lncRNA MCF2L-AS1 (MCF2 cell-line-derived transforming sequence-like 2 antisense RNA 1) and cyclin D1 (CCND1) [9]. The interactions of MCF2L-AS1 and CCND1 mediated by HuR/ELAVL1 revealed a new potential mechanism for NSCLC progression and development of resistance to the widely used drug gefitinib. Cyclin D1 (CCND1) is an oncoprotein involved in the regulation of the cell cycle and the transition from the G1 phase to the S phase. Both the lncRNA MCF2L-AS1 and CCND1 were overexpressed in NSCLC cell lines. In addition, MCF2L-AS1 was upregulated by the transcription factor E2F1, which binds to the MCF2L-AS1 promoter region, as predicted by JASPAR data [9]. The direct interaction between them was confirmed by luciferase and ChIP assays [9], while the direct interaction between MCF2L-AS1, CCND1, and HuR/ELAVL1 was validated using RNA pull-down and RIP assays [9]. These experiments showed that MCF2L-AS1, activated by the binding of transcription factor E2F1 to the promoter region in complex with HuR/ELAVL1, interacts with the CCND1 mRNA, increasing its stability, driving NSCLC progression, and inducing patient resistance to gefitinib (Table 1).
An intriguing mechanism reducing NSCLC stemness was found for the suppressor lncRNA FENDRR (FOXF1 adjacent non-coding developmental regulatory RNA), also known as FOXF1-AS1. It is mediated by the inhibition of oncogenic multidrug resistance gene 1 (MDR1) mRNA and RBP HuR [8]. FENDRR lncRNA expression is decreased in NSCLC tissues and cells, and its overexpression abates the stemness of NSCLC cells. The latter was confirmed by stemness marker (CD34 and CD133) expression analysis and the capacity of cells for spheroid formation. Direct binding of lncRNA FENDRR to MDR1 3’UTR was shown using such methods as the luciferase reporter and RIP assays [8]. MDR1 mRNA stability was measured in NSCLC cells in the presence of actinomycin D with or without FENDRR overexpression. MDR1 mRNA stability was significantly decreased upon FENDRR overexpression. Thus, lncRNA FENDRR directly binds to the MDR1 3′UTR and reduces MDR1 mRNA stability [8].
RBP HuR can promote the stabilization of target transcripts and bind to mRNA 3′UTR with AU-rich elements. Direct interaction of HuR with the MDR1 3’UTR was validated using luciferase reporter and RIP assays [8]. In addition, RBP HuR increased the expression and stability of MDR1 mRNA. Moreover, lncRNA FENDRR was shown to compete with HuR for binding to MDR1 3′-UTR, and its overexpression could partially prevent HuR binding to MDR1 3’UTR. Thus, lncRNA FENDRR and RBP HuR, two crucial epigenetic regulators capable of binding to 3’UTR of MDR1, were demonstrated to exert opposite effects and compete with each other [8]. FENDRR lncRNA suppressed NSCLC cell stemness by inhibiting MDR1, whereas RBP HuR, which competed with FENDRR, played the opposite role (Table 1).
HuR RBP is involved in elevating the level and stability of PD-L1 (programmed cell death 1 ligand 1) and USP8 (ubiquitin-specific processing protease 8) mRNA by lncRNA SNHG12 (small nucleolar RNA host gene 12). As PD-L1 stabilization contributes to the evasion of the immune response in NSCLC, targeting this pathway can be promising for NSCLC immunotherapy [10]. The recently discovered immune checkpoint (PD-L1/PD-1) blocks production of antibodies and cytokines in cancer, thus impairing immune cell activation and reducing the immune response towards tumor cells. HuR at the post-transcriptional level increased the level and stability of USP8 mRNA and facilitated translation of USP8 protein, promoting the proliferation, migration, and invasion of NSCLC cells. USP8 is a deubiquitinase that can inhibit ubiquitin-dependent degradation and increase the stability of oncoproteins. SNHG12 is highly expressed in NSCLC tissues and cells [10]. An increase in the SNHG12 lncRNA expression was shown to be associated with a shorter overall survival of patients with NSCLC (Kaplan–Meier curves). The half-life of USP8 and PD-L1 mRNAs in NSCLC cells was assessed by RT-qPCR. ChIP assay confirmed the interaction between USP8 and PD-L1 proteins. USP8 was shown to stabilize PD-L1 through deubiquitination. Direct binding of SNHG12 to HuR, as well as binding of PD-L1 and USP8 to HuR, was predicted using RNA–protein interaction prediction (RPISeq) (http://pridb.gdcb.iastate.edu/RPISeq/, accessed on 19 June 2023) and validated by RIP assay [10]. SNHG12 was shown to elevate the expression and stability of PD-L1 and USP8 mRNA as well as the level of translation of the USP8 protein due to its binding to HuR. Deubiquitination of PD-L1 suppresses immune CD8+ T cells and contributes to the escape of NSCLC from the immune response (Table 1).
3. Heterogeneous Nuclear Ribonucleoproteins with RBP Function
Heterogeneous nuclear ribonucleoproteins (hnRNPs) are involved in many processes, including alternative splicing, transcription and translation regulation, and mRNA stabilization [23]. Possessing RBP properties, hnRNPs can mediate the regulatory effect of lncRNA on target mRNA or play a supportive role. In NSCLC, lncRNAs function together with three heterogeneous nuclear ribonucleoproteins: hnRNPD (or AUF1), hnRNPK, and hnRNPU.
AUF1 (AU-rich element RNA binding/degradation factor 1) is a heterogeneous nuclear ribonucleoprotein (hnRNPD) recognized as a classical RBP. AUF1 has a positive effect on the immune response towards NSCLC and demonstrates tumor suppressor properties. AUF1 functions as an antagonist of the oncogenic lncRNA SChLAP1 preventing SChLAP1-dependent stabilization of PDL-1 mRNA which allows NSCLC cells to evade the immune response [11]. LncRNA SChLAP1 (SWI/SNF complex antagonist associated with prostate cancer 1) is upregulated in NSCLC and promotes NSCLC cell proliferation, migration, and invasion [11], while its knockdown represses tumor growth and metastasis in vivo. SChLAP1 also facilitates immune evasion by enhancing PD-L1 mRNA stability and might inhibit AUF1 as a negative regulator of mRNA stability. SChLAP1 binding to AUF1 was predicted from the RNA-Protein Interaction Prediction (RPISeq) database (http://pridb.gdcb.iastate.edu/RPISeq/, accessed on 19 June 2023) and validated using the RIP and RNA pull-down assays [11]. SChLAP1 binding to AUF1 was shown to decrease AUF1 level and prevent AUF1 from suppressing PD-L1 mRNA stability. SChLAP1 overexpression decreased AUF1 enrichment in the 3’UTR region of PD-L1. Of note, PD-L1 upregulation induced by SChLAP1 overexpression was abolished upon AUF1 overexpression [11]. As a result, SChLAP1 overexpression elevated the levels of PD-L1 mRNA and protein. Thus, the oncogenic lncRNA SChLAP1 and RBP AUF1 compete for effects on the expression and stability of PD-L1 at the post-transcriptional level. PD-L1 is usually highly expressed on the surface of tumor cells repressing CD8+ T cell function. In summary, SChLAP1 binds to AUF1, reducing the interaction between AUF1 and the PD-L1 3’UTR and thus increasing PD-L1 mRNA stability and expression, which in turn represses CD8+ T cell function and facilitates tumor cell immune escape (Table 1).
Heterogeneous nuclear ribonucleoprotein hnRNPK is a key RNA-binding protein with oncogenic functions. In NSCLC, it stimulates the transcription of its target, the c-MYC oncogene, by participating in the binding of RNA polymerase II (RNA Pol II) to c-MYC [12]. The lncRNA DIO3OS (DIO3 opposite strand upstream RNA, or antisense lncRNA transcribed from the DIO3 (type 3 iodothyronine deiodinase) imprinted locus) in NSCLC directly interacts with hnRNPK, repressing hnRNPK binding to MYC DNA and mRNA, and inhibits MYC transcription and translation [12]. Furthermore, the lncRNA DIO3OS can suppress CDC25A, a downstream MYC target (Table 1). Ectopic expression of DIO3OS lncRNA suppresses NSCLC tumorigenesis and metastasis in vitro and in vivo. However, all these effects can be abolished by methylation of the CpG-456 dinucleotide gene encoding DIO3OS lncRNA, which can occur in NSCLC. The direct interactions between DIO3OS lncRNA, hnRNPK, c-MYC, CDC25A, and RNA Pol II in NSCLC were confirmed by complex methods such as dual-luciferase reporter assay, RNA pull-down, RIP and ChIP assays, and Western blotting [12].
RBP hnRNPU, which exhibits a suppressor function, mediates the interactions of suppressor LIMD1 (LIM domains-containing 1) mRNA with suppressor lncRNA LIMD1-AS1 (LIMD1 antisense RNA 1), suppressing NSCLC growth in vitro and in vivo [13] (Table 1). Moreover, LIMD1-AS1 interaction with hnRNPU stabilized LIMD1 mRNA. Unlike hnRNPK [12] and hnRNPD AUF1 [11], which prevented lncRNA from interacting with mRNA via competitive binding, hnRNPU promoted the interaction of lncRNA LIMD1-AS1 with the mRNA target LIMD1. Their direct binding was confirmed using RIP and pull-down assays in [13].
4. Other RBPs (FBL, EIF4A3, UPF1, WDR5, YTHDF1/2/3) as Mediators of lncRNAs
FBL, EIF4A3, UPF1, WDR5, and YTHDF1/2/3 also act as RBP mediators of lncRNA action on protein-coding genes in NSCLC.
FBL was found to be an RBP mediator in the interaction between FAM83A antisense transcript 1 (FAM83A-AS1) and FAM83A pre-mRNA (family with sequence similarity 83 member A). Both FAM83A and FAM83A-AS1 are typical pro-tumor genes and are overexpressed in NSCLC, they activate cell migration and metastasis in vitro, and their levels correlate with OS and PFS [14]. RNase protection assay and RT-qPCR were used to detect duplex formation within the FAM83A pre-mRNA and FAM83A-AS1 overlapping region identified between exon 3 and exon 4 of the FAM83A gene. FAM83A-AS1 was shown to enhance FAM83A stability due to formation of the RNA–RNA duplex. The RNA pull-down and RIP assays and RT-qPCR showed the interaction of FBL RBP both with FAM83A and FAM83A-AS1 [14]. This indicates that an increase in FAM83A mRNA stability occurs not only due to the RNA–RNA interaction but also due to a triple RNP complex formed with the FBL protein [14]. Moreover, FAM83A-AS1 promotes NSCLC progression via ERK signaling pathways and metastasis by increasing FAM83A expression through FAM83A-AS1 to FAM83A pre-mRNA binding by forming an RNA/mRNA heteroduplex. RBP FBL binds the duplex enhancing FAM83A mRNA stability (Table 1).
Various antisense genes are involved in regulating their neighboring genes. Another example of RBP-dependent regulation of a protein-coding gene by an adjacent complementary antisense transcript is the TM4SF19 gene (transmembrane 4 L six family member 19). It is regulated by the conservative lncRNA TM4SF19-AS1 encoded by the TM4SF19 gene cluster, located at chromosome 3q29, and depends on WDR5 (WD repeat-containing protein 5) [17]. TM4SF19-AS1, like TM4SF19, was both upregulated in cells and patients’ tissues of lung squamous cell carcinoma (LSCC). Like TM4SF19, TM4SF19-AS1 can promote LSCC cell proliferation and adhesion. Moreover, there was a markedly positive correlation between TM4SF19-AS1 and TM4SF19 levels [17].
Furthermore, TM4SF19-AS1 activated TM4SF19 via WDR5 binding and inducing demethylation of the TM4SF19 promoter region. WDR5 was shown to exert two functions. WDR5 is a key subunit of the chromatin remodeling complex MLL1 (mixed-lineage leukemia 1), capable of enforcing active chromatin. It is also an RBP subunit, as it contains an RNA-binding pocket which allows lncRNA binding and regulates their target genes [24]. Moreover, WDR5 can be introduced into the promoter region of its target gene by lncRNAs to demethylate the target gene promoter and activate target gene expression. Similar results were obtained for lncRNA TM4SF19-AS1, which interacted with WDR5 and delivered it to the TM4SF19 promoter causing subsequent promoter demethylation and TM4SF19 upregulation [17].
This mechanism was validated by several different approaches. First, FISH showed that lncRNA TM4SF19-AS1 is localized in the nucleus and can interact with DNA. The direct interaction of WDR5 with the TM4SF19 gene promoter was validated using ChIP with TM4SF19-specific antibodies. In addition, the methylation status of the TM4SF19 promoter region was determined using a methylation-specific PCR (MSP) assay. TM4SF19 promoter methylation was reduced in LSCC cell lines, but its methylation level was elevated after si-TM4SF19-AS1 treatment. WDR5 binding to TM4SF19-AS1 was validated by RIP with WDR5-specific antibodies [17].
In summary, there is substantial evidence that in LSCC, the interaction between TM4SF19-AS1 and WDR5 reduced the TM4SF19 methylation leading to TM4SF19 upregulation (Table 1). These data represent a rare example of the interaction of lncRNAs with DNA, i.e., with the promoter of a protein-coding gene mediated by RBP.
Eukaryotic translation initiation factor 4A3 (EIF4A3) is a well-characterized RBP that regulates the expression of non-coding RNAs in tumors [25]. EIF4A3, an RNA helicase and a core component of the exon junction complex, plays crucial roles in splicing and is activated in a variety of cancers and involved in autophagy regulation [25][26]. EIF4A3 is also involved in the regulation of angiogenesis [15]. Thus, EIF4A3 was shown to contribute to LINC00667-dependent stimulation of vascular endothelial cell proliferation and migration by increasing the stability of vascular endothelial growth factor A (VEGFA). LINC00667 is overexpressed in NSCLC and, according to the dual luciferase reporter assay, it cannot bind and regulate VEGFA promoter activity; on the contrary, it can regulate the expression of VEGFA at the post-transcriptional level [15]. StarBase v3.0 scanning revealed 71 RBPs capable of binding to both LINC00667 and VEGFA mRNA, five (ILF3, MOV10, EIF4A3, ADAR, and IGF2BP2) being overexpressed in NSCLC. Knockdown experiments demonstrated that among these five RBPs, only EIF4A3 could increase the mRNA and protein levels of VEGFA [15] (Table 1). Experiments with actinomycin D showed the ability of EIF4A3 to increase the half-life of VEGFA mRNA in NSCLC. Furthermore, direct binding of EIF4A3 protein to VEGFA mRNA and lncRNA LINC00667 was validated using RIP and RNA pull-down assays. In summary, LINC00667 in the RNP complex containing EIF4A3 RBP and VEGFA mRNA can promote the stabilization of VEGFA mRNA, proliferation, migration, and neoangiogenesis in NSCLC [15].
Most relapses after surgery in patients with NSCLC indicate the presence of persisting cancer stem cells (CSCs). CSC progression is associated with the Hippo pathway [27][28]. The upregulated Hippo pathway kinase LATS1/2 (large tumor suppressor kinase 1, 2) can phosphorylate and inactivate YAP and TAZ. Several other lncRNAs contribute to maintaining the stemness in NSCLC. For instance, the antisense lncRNA MACC1-AS1 is a homolog of the last intron of the metastasis associated in colon cancer 1 (MACC1) gene. MACC1-AS1 exhibits carcinogenic properties and is upregulated in NSCLC [16]. Its overexpression inhibited YAP and TAZ, inactivated the Hippo pathway, and enhanced the stemness of NSCLC cells via downregulating the expression of LATS1/2 kinase at the mRNA level.
Up-frameshift protein 1 (UPF1) is an evolutionarily conserved protein with RNA/DNA-dependent ATPase and RNA helicase activity. UPF1 can reduce mRNA stability; in particular, it can repress the stability of suppressive LATS1/2 kinase mRNA. Direct binding of UPF1 to both MACC1-AS1 and LATS1/2 mRNA was demonstrated by RIP analysis using the RNA-Binding Protein Immunoprecipitation Kit [16]. Overall, MACC1-AS1 in complex with UPF1 RBP can drive NSCLC cell stemness via inhibiting the Hippo pathway LATS1/2 kinase (Table 1).
Many known RBP m6A readers contain an YTH domain that specifically recognizes m6A over A. They belong to the YTHDF1/2/3 family and are predominantly localized in the cytoplasm [29]. m6A-modified lncRNA DLGAP1 (disks large-associated protein 1) antisense RNA 2 (DLGAP1-AS2) was shown to play an important role in NSCLC [18]. METTL3 m6A-methyltransferase enhanced its stability via m6A transfer to DLGAP1-AS2. At the same time, YTHDF1, the m6A-reader RBP that functions as a part of the RNP complex, was shown to transfer m6A to c-Myc mRNA, increasing its stability [18]. This mechanism mediates DLGAP1-AS2-dependent oncogenesis and NSCLC progression, stimulation of c-Myc-dependent aerobic glycolysis, and deteriorated prognosis of patients (Table 1).
5. LncRNAs Mediated by Both miRNA and RBP
It is worth noting that there are a number of lncRNAs that can function by both the ceRNA mechanism and alternative mechanisms. Figure 1a–c shows three lncRNAs (DLGAP1-AS2, MNX1-AS1, and SNHG12) that can act via both miRNA and RBP. For the extensively studied MALAT1, ATG12 upregulation was shown to involve RBP IGF2BP2 as well as different axes according to the ceRNA model (Figure 1d). Moreover, IGF2BP2 stabilizes MALAT1 due to m6A methylation.
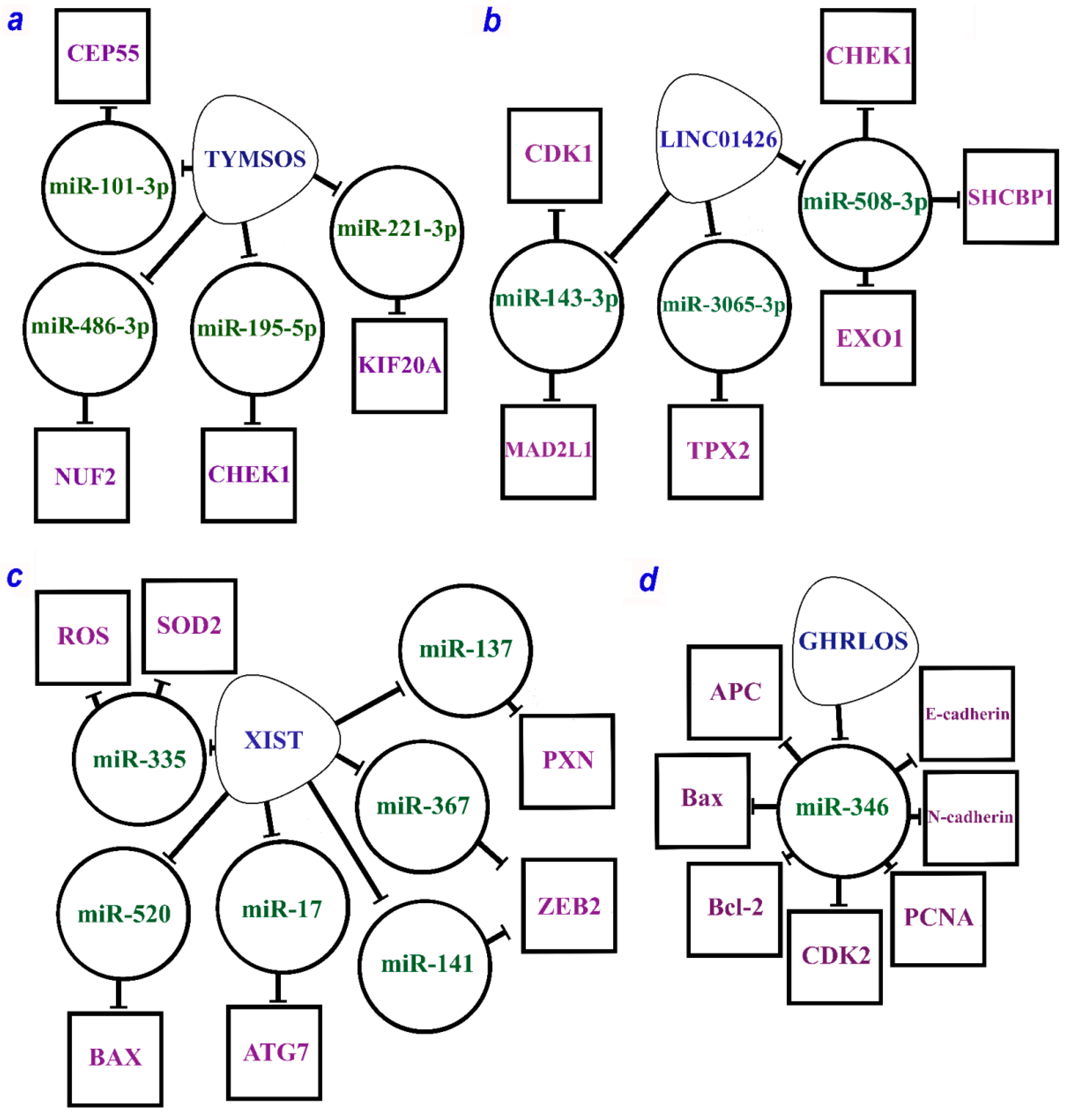
Figure 1. The multiple mechanisms of lncRNAs (a) DLGAP1-AS2 (DLGAP1 antisense RNA 2), (b) MNX1-AS1 (MNX1 antisense RNA 1), (c) SNHG12 (small nucleolar RNA host gene 12), and (d) MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) by ceRNA model and through the RBP (according to the following works [3][6][10][18][30][31][32][33][34][35]). LncRNAs are within the soft triangles; miRNAs are within the circles; mRNA/proteins are within the squares; blunt arrows indicate inhibitory interactions; a straight arrow indicates activation of targets.
References
- Pereira, B.; Billaud, M.; Almeida, R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 2017, 3, 506–528.
- Zhu, L.; Liu, Y.; Tang, H.; Wang, P. FOXP3 activated-LINC01232 accelerates the stemness of non-small cell lung carcinoma by activating TGF-beta signaling pathway and recruiting IGF2BP2 to stabilize TGFBR1. Exp. Cell Res. 2022, 413, 113024.
- Zhu, Q.; Zhang, C.; Qu, T.; Lu, X.; He, X.; Li, W.; Yin, D.; Han, L.; Guo, R.; Zhang, E. MNX1-AS1 Promotes Phase Separation of IGF2BP1 to Drive c-Myc-Mediated Cell-Cycle Progression and Proliferation in Lung Cancer. Cancer Res. 2022, 82, 4340–4358.
- Yang, J.; Qian, X.; Qiu, Q.; Xu, L.; Pan, M.; Li, J.; Ren, J.; Lu, B.; Qiu, T.; Chen, E.; et al. LCAT1 is an oncogenic LncRNA by stabilizing the IGF2BP2-CDC6 axis. Cell Death Dis. 2022, 13, 877.
- Brownmiller, T.; Juric, J.A.; Ivey, A.D.; Harvey, B.M.; Westemeier, E.S.; Winters, M.T.; Stevens, A.M.; Stanley, A.N.; Hayes, K.E.; Sprowls, S.A.; et al. Y Chromosome LncRNA Are Involved in Radiation Response of Male Non-Small Cell Lung Cancer Cells. Cancer Res. 2020, 80, 4046–4057.
- Han, L.; Lei, G.; Chen, Z.; Zhang, Y.; Huang, C.; Chen, W. IGF2BP2 Regulates MALAT1 by Serving as an N6-Methyladenosine Reader to Promote NSCLC Proliferation. Front. Mol. Biosci. 2021, 8, 780089.
- Jiao, P.F.; Tang, P.J.; Chu, D.; Li, Y.M.; Xu, W.H.; Ren, G.F. Long Non-Coding RNA THOR Depletion Inhibits Human Non-Small Cell Lung Cancer Cell Growth. Front. Oncol. 2021, 11, 756148.
- Gong, F.; Dong, D.; Zhang, T.; Xu, W. Long non-coding RNA FENDRR attenuates the stemness of non-small cell lung cancer cells via decreasing multidrug resistance gene 1 (MDR1) expression through competitively binding with RNA binding protein HuR. Eur. J. Pharmacol. 2019, 853, 345–352.
- Shan, K.Z.; Yang, S.F.; Deng, Y.J.; Yue, P.Y.; Du, Z.Q. E2F1-induced long non-coding RNA MCF2L-AS1 modulates Cyclin D1 mRNA stability through ELAVL1 to induce Gefitinib resistance in non-small cell lung cancer. Acta Biochim. Pol. 2022, 69, 795–804.
- Huang, Y.; Xia, L.; Tan, X.; Zhang, J.; Zeng, W.; Tan, B.; Yu, X.; Fang, W.; Yang, Z. Molecular mechanism of lncRNA SNHG12 in immune escape of non-small cell lung cancer through the HuR/PD-L1/USP8 axis. Cell Mol. Biol. Lett. 2022, 27, 43.
- Du, Z.; Niu, S.; Wang, J.; Wu, J.; Li, S.; Yi, X. SChLAP1 contributes to non-small cell lung cancer cell progression and immune evasion through regulating the AUF1/PD-L1 axis. Autoimmunity 2021, 54, 225–233.
- Zhang, M.; Wu, J.; Zhong, W.; Zhao, Z.; He, W. DNA-methylation-induced silencing of DIO3OS drives non-small cell lung cancer progression via activating hnRNPK-MYC-CDC25A axis. Mol. Ther. Oncolytics 2021, 23, 205–219.
- Pan, J.; Tang, Y.; Liu, S.; Li, L.; Yu, B.; Lu, Y.; Wang, Y. LIMD1-AS1 suppressed non-small cell lung cancer progression through stabilizing LIMD1 mRNA via hnRNP U. Cancer Med. 2020, 9, 3829–3839.
- Wang, W.; Zhao, Z.; Xu, C.; Li, C.; Ding, C.; Chen, J.; Chen, T.; Zhao, J. LncRNA FAM83A-AS1 promotes lung adenocarcinoma progression by enhancing the pre-mRNA stability of FAM83A. Thorac. Cancer 2021, 12, 1495–1502.
- Yang, H.; Yang, W.; Dai, W.; Ma, Y.; Zhang, G. LINC00667 promotes the proliferation, migration, and pathological angiogenesis in non-small cell lung cancer through stabilizing VEGFA by EIF4A3. Cell Biol. Int. 2020, 44, 1671–1680.
- Wang, X.; Yu, X.; Wei, W.; Liu, Y. Long noncoding RNA MACC1-AS1 promotes the stemness of nonsmall cell lung cancer cells through promoting UPF1-mediated destabilization of LATS1/2. Environ. Toxicol. 2020, 35, 998–1006.
- Luo, M.; Xie, L.; Su, Y.; Zhang, K.; Liang, R.; Ma, Z.; Li, Y. TM4SF19-AS1 facilitates the proliferation of lung squamous cell carcinoma by recruiting WDR5 to mediate TM4SF19. Mol. Cell Probes 2022, 65, 101849.
- Zhang, Q.; Zhang, Y.; Chen, H.; Sun, L.N.; Zhang, B.; Yue, D.S.; Wang, C.L.; Zhang, Z.F. METTL3-induced DLGAP1-AS2 promotes non-small cell lung cancer tumorigenesis through m6A/c-Myc-dependent aerobic glycolysis. Cell Cycle 2022, 21, 2602–2614.
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326.
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624.
- Brennan, C.M.; Steitz, J.A. HuR and mRNA stability. Cell Mol. Life Sci. CMLS 2001, 58, 266–277.
- Lebedeva, S.; Jens, M.; Theil, K.; Schwanhausser, B.; Selbach, M.; Landthaler, M.; Rajewsky, N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell 2011, 43, 340–352.
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016, 135, 851–867.
- Yang, Y.W.; Flynn, R.A.; Chen, Y.; Qu, K.; Wan, B.; Wang, K.C.; Lei, M.; Chang, H.Y. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife 2014, 3, e02046.
- Zhu, Y.; Ren, C.; Yang, L. Effect of eukaryotic translation initiation factor 4A3 in malignant tumors. Oncol. Lett. 2021, 21, 358.
- Sakellariou, D.; Frankel, L.B. EIF4A3: A gatekeeper of autophagy. Autophagy 2021, 17, 4504–4505.
- Tang, Y.; Feinberg, T.; Keller, E.T.; Li, X.Y.; Weiss, S.J. Snail/Slug binding interactions with YAP/TAZ control skeletal stem cell self-renewal and differentiation. Nat. Cell Biol. 2016, 18, 917–929.
- Wang, L.; Zhang, Z.; Yu, X.; Huang, X.; Liu, Z.; Chai, Y.; Yang, L.; Wang, Q.; Li, M.; Zhao, J.; et al. Unbalanced YAP-SOX9 circuit drives stemness and malignant progression in esophageal squamous cell carcinoma. Oncogene 2019, 38, 2042–2055.
- Patil, D.P.; Pickering, B.F.; Jaffrey, S.R. Reading m(6)A in the Transcriptome: M(6)A-Binding Proteins. Trends Cell Biol. 2018, 28, 113–127.
- Wang, L.; Tang, L.; Ge, T.; Zhu, F.; Liu, D.; Guo, H.; Qian, P.; Xu, N. LncRNA DLGAP1-AS2 regulates miR-503/cyclin D1 to promote cell proliferation in non-small cell lung cancer. BMC Pulm. Med. 2021, 21, 277.
- Rong, F.; Liu, L.; Zou, C.; Zeng, J.; Xu, Y. MALAT1 Promotes Cell Tumorigenicity through Regulating miR-515-5p/EEF2 Axis in Non-Small Cell Lung Cancer. Cancer Manag. Res. 2020, 12, 7691–7701.
- Wang, Y.; Zhang, Q. Long Noncoding RNA MALAT1 Knockdown Inhibits Proliferation, Migration, and Invasion and Promotes Apoptosis in Non-Small-Cell Lung Cancer Cells through Regulating miR-515-3p/TRIM65 Axis. Cancer Biother. Radiopharm. 2020; ahead of print.
- Yu, W.; Ding, J.; He, M.; Chen, Y.; Wang, R.; Han, Z.; Xing, E.Z.; Zhang, C.; Yeh, S. Estrogen receptor beta promotes the vasculogenic mimicry (VM) and cell invasion via altering the lncRNA-MALAT1/miR-145-5p/NEDD9 signals in lung cancer. Oncogene 2019, 38, 1225–1238.
- Li, S.; Mei, Z.; Hu, H.B.; Zhang, X. The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. J. Cell Physiol. 2018, 233, 6679–6688.
- Tan, D.; Wang, S.; Zhang, P.; Peng, C.; Wu, T. LncRNA SNHG12 Decreases Non-Small Cell Lung Cancer Cell Sensitivity to Cisplatin by Repressing miR-525-5p and Promoting XIAP. Ann. Clin. Lab. Sci. 2023, 53, 64–75.
More
