Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Francesco Zito and Version 3 by Rita Xu.
The ability to simulate gene expression and infer gene regulatory networks has vast potential applications in various fields, including medicine, agriculture, and environmental science. Machine learning approaches to simulate gene expression and infer gene regulatory networks have gained significant attention as a promising area of research.
- reverse engineering
- gene regulatory network
- machine learning
1. Introduction
Understanding the intrinsic relationship between genes with the aim of treating known diseases is currently one of the great challenges in genetics [1]. Although this topic may seem to be only a biological problem, it actually involves many areas of computer science. Due to the complexity of this problem, traditional mathematical methods such as Ordinary Differential Equations (ODEs), which rely on estimates of gene expression levels over time through a continuous model, may be inaccurate for a larger number of genes and require high-quality data to create an acceptable model [2]. Machine-learning-based techniques have emerged as a promising approach for gene regulatory network inference, outperforming other methods based on mutual information [3]. These techniques can be broadly classified into two categories. The first category involves using observations to create a model that approximates the real system, which is then used to construct a complex network that identifies the regulatory genes for other genes, known as the gene regulatory network [4]. The second category involves the direct creation of a gene regulatory network through observations, without the need to estimate a model representing the dynamics of gene expression [3][5][3,5].
Related Work
Before the spread of machine learning in the field of genetics, Boolean networks were generally used to describe gene regulatory networks. All biological components can be described by binary states and their interactions by Boolean functions [6][9]. Boolean networks are relatively simple to implement, but their implementation requires noise-free, discrete data, which can be difficult to obtain when working with real-world data [7][10].
In recent years, several methods to extract a gene regulatory network have been presented. In [8][11], the authors divided the methods for inferring a gene regulatory network from gene expression data into three main groups: (i) model-based methods; (ii) information-theory-based methods; and (iii) machine learning methods. Some experimental tests have shown that machine learning methods can obtain a high accuracy in predicting gene interactions [9][12]. One approach to inferring a gene regulatory network from a gene expression dataset is to use differential equations. This requires a mathematical model of the changes in gene expression over time using ordinary differential equations, which can provide insight into the underlying dynamics of the system. By analyzing the behavior of these equations, one can gain a better understanding of how genes interact and regulate each other within the network [10][13]. The difficulty in such an approach is, clearly, to build a differential equation model from data. To this end, several methods have been proposed in the literature. An example can be found in [11][14], where a metaheuristic was used to find the parameters of an S-system model describing the dynamics of gene expression. Another example can be found in [12][15], where a complex-valued ordinary differential equation model was created using genetic programming. In addition, it is possible to directly predict the interaction between genes using a gene expression dataset. One method used in this field is GENIE3 presented in [13][16] and its improvement called DynamicGENIE3 presented in [14][17]. An improvement on the previously cited method in this category can be found in [15][18], where different inference methods are combined to increase the accuracy of the resulting gene regulatory network.
Rather than using a specific strategy to predict each arc of a gene regulatory network, an alternative approach involves the construction of a comprehensive network that assumes all possible interactions between genes, represented as a strongly connected graph, and subsequently applying a pruning strategy to eliminate non-corresponding arcs. One example of such a method, which employs an information–theoretic algorithm, is described in [16][19].
2. SimulBate Gene Expression anckground Infer Gene Regulatory Networks
The process by which the instructions in our DNA are transformed into a functioning product, such as a protein, is known as gene expression [17][20]. Gene expression allows a cell to respond to changes in its environment. The regulation of gene expression (or just gene regulation) is a very complex process that takes into account several biological factors to respond, for example, to environmental stimuli or to adapt to new food sources [18][19][21,22]. Gene regulation involves a variety of mechanisms used by cells to increase or decrease the production of certain gene products. Thus, it functions like an on/off switch that regulates the amount of proteins produced. Considering the huge amount of gene products that are present in a multicellular organism, the regulatory mechanisms are represented in a directed graph, called the regulatory network, to help better understand the regulatory mechanisms. A regulatory network reveals the interactions between genes, proteins, mRNAs, and cellular processes and provides important information about the development of diseases [20][23]. Knowledge of a regulatory network for an entire organism or for a small group of genes is crucial for a full understanding of the life process of an organism and how gene products interact with each other [21][24]. Once this is clear, it is possible to send external chemical signals to inhibit a gene that could be dangerous to the life of an organism, such as the development of a cancer cell or a genetic disease [22][25].2.1. Gene Regulatory Network
A gene regulatory network is a directed graph where the nodes represent genes, and the directed arcs model the interactions between the genes [23][26]. Specifically, a Gene Regulatory Network (GRN) represents the regulatory process of gene expression in an organism. An arc between two nodes, i.e., genes, mainly provides information about the regulatory process. In the context of inferring gene regulatory networks, the presence of a direct arc from gene 𝐺𝑖 to gene 𝐺𝑗 indicates that 𝐺𝑖 is a regulatory gene, also known as a regulator [24][27]. This implies that any alteration in the expression of 𝐺𝑖 will have a consequential impact on the expression of 𝐺𝑗, according to the principle of cause and effect. In other words, the regulatory gene 𝐺𝑖 is capable of influencing the expression of its target gene 𝐺𝑗, thereby establishing a cause-and-effect relationship between the two genes.A gene regulatory network can, therefore, combine more-detailed regulatory information. In fact, a regulatory gene controls the expression of its associated genes in a positive or negative way. When the expression level of the regulator reaches a threshold, another gene can be activated or inhibited based on that level [25][28]. This results in a change in the expression level of the regulated gene: if the gene expression decreases, the gene is inhibited; otherwise, it is activated. Figure 1 shows an example of a gene regulatory network. As can be seen, there are two types of arcs in a gene regulatory network: activation arcs and inhibition arcs.
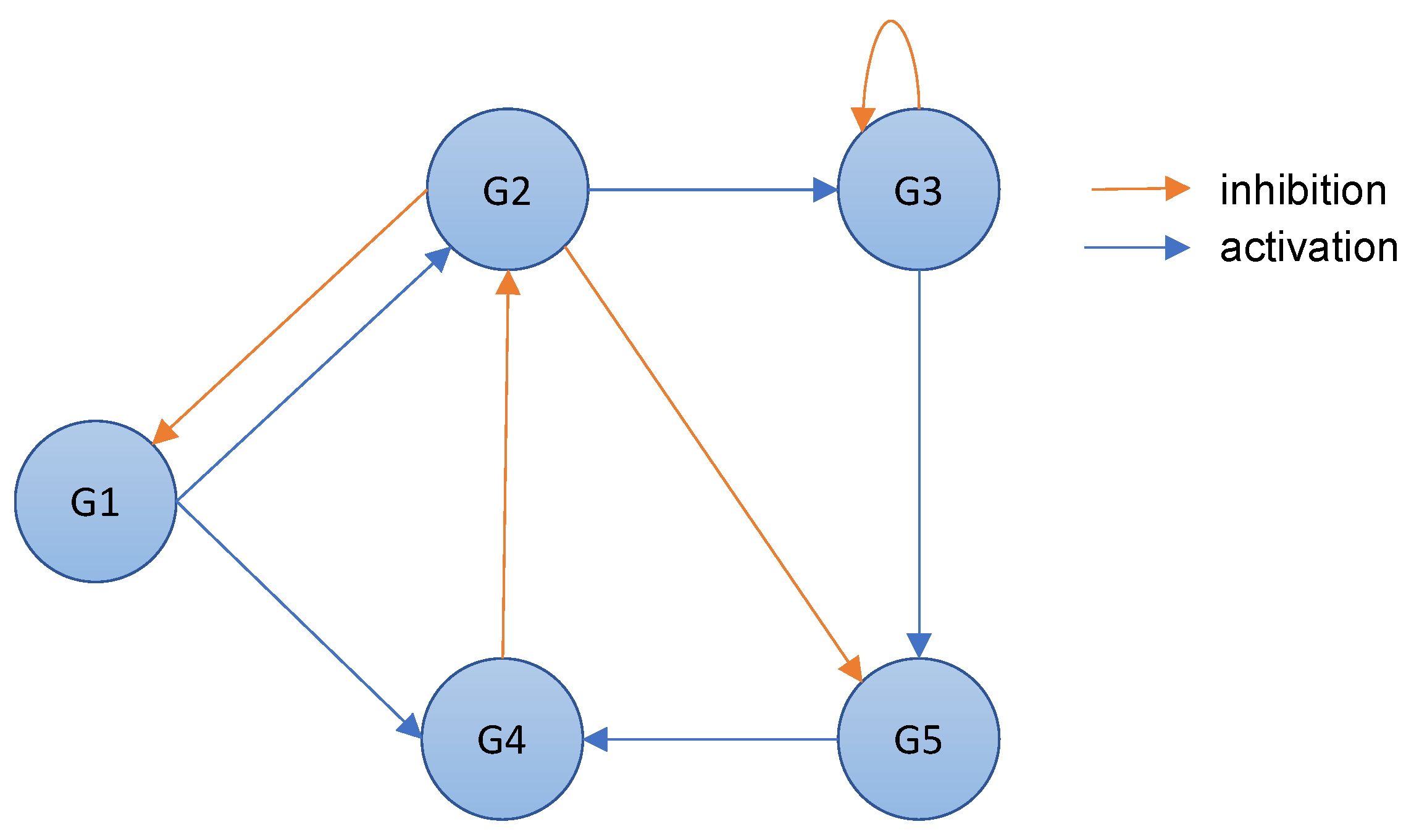
Figure 1. An example of a gene regulatory network that includes gene regulation information.
2.2. Inferring a Gene Regulatory Network
The process of inferring a gene regulatory network for a cellular organism can be divided into four distinct phases, which rwesearchers label: observation; modeling; inference; and validation. The whole process is shown in Figure 2.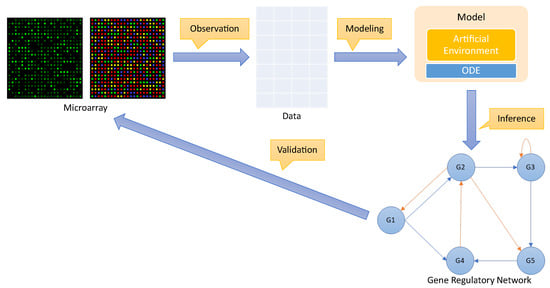
Figure 2. Process to infer a gene regulatory network.
-
Observation: The first step is to observe how the gene expression of a group of genes responds to external perturbations in a real organism. This can be performed using various strategies, such as microarray technology [26]. The level of gene expression for each gene is recorded over time to create a time-series dataset containing gene expression for the genes under observation. Typically, such a dataset is represented as a matrix , where N is the number of genes and M is the number of observations for each gene over time.
-
Modeling: The gene expression time-series dataset is used to train a model that can be based on differential equations [27] or design an artificial environmental setting.
-
Inference: The model created in the previous phase is used to make predictions about the relationships between genes in order to discover regulatory genes. This information can, therefore, be used to draw a complex network, i.e., a gene regulatory network, showing these relationships.
-
Validation: Finally, to validate the accuracy of a predicted gene regulatory network, it is essential to compare it with the target network. However, this comparison can only be performed on an artificial dataset where the gene regulatory network is known beforehand. In a real organism, researchers do not have access to a gene regulatory network, and therefore, the validation of the predicted gene regulatory network must be performed empirically and in the field.
-
Observation: The first step is to observe how the gene expression of a group of genes responds to external perturbations in a real organism. This can be performed using various strategies, such as microarray technology [29]. The level of gene expression for each gene is recorded over time to create a time-series dataset containing gene expression for the genes under observation. Typically, such a dataset is represented as a matrix , where N is the number of genes and M is the number of observations for each gene over time.
-
Modeling: The gene expression time-series dataset is used to train a model that can be based on differential equations [30] or design an artificial environmental setting.
-
Inference: The model created in the previous phase is used to make predictions about the relationships between genes in order to discover regulatory genes. This information can, therefore, be used to draw a complex network, i.e., a gene regulatory network, showing these relationships.
-
Validation: Finally, to validate the accuracy of a predicted gene regulatory network, it is essential to compare it with the target network. However, this comparison can only be performed on an artificial dataset where the gene regulatory network is known beforehand. In a real organism, we do not have access to a gene regulatory network, and therefore, the validation of the predicted gene regulatory network must be performed empirically and in the field.
