Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Zainul Abideen and Version 2 by Rita Xu.
The relationship between plants and nanoparticles (NPs) has been the subject of extensive research interest. Nanoparticles and their interactive effects on plants have raised concern regarding their harmful aspects. There are complex mechanisms evolved in plants for controlling the uptake, accumulation, and mobilization of nanoparticles that need to be discussed further, especially in the context of proteomics and genetic level.
- nanotechnology
- omics-based studies
- plant–metal interaction
1. Introduction
Nanotechnology is a rapidly emerging and multifaceted science with great potential and applications. The concern of the scientific community regarding its impact on biota has grown along with the technology’s rapid growth [1][2][3][1,2,3]. Given that manufactured nanomaterials (MNMs) or engineered nanoparticles (ENPs) play a significant role in this rapidly emerging science, the remarkable physicochemical properties of manufactured nanoscale materials (ENMs) are studied in precision agriculture to (i) increase nutritional quality and crop productivity [4], (ii) apply the controlled release and targeted delivery of agrochemicals [5][6][5,6], (iii) protect crops against pathogens [7][8][7,8], (iv) enhance resilience to environmental stress [9][10][9,10], and (v) monitor soil quality and detect stressors using sensitive nanosensors [11][12][11,12]. In addition to their benefits to the targeted crop, it is also possible to significantly reduce unwarranted exposure to nontargeted organisms and leaching into environmental waters. The concern of the scientific community regarding its impact on biota has grown along with the technology’s significant role in a variety of consumer goods, including biomedicine, cosmetics, and many others, and their massive atmospheric release is inevitable. Plant communities are particularly exposed to MNMs due to their restricted distribution [13][14][15][13,14,15]. There is a lot of literature on nanoparticles’ (NPs’) beneficial as well as phytotoxic implications for plant communities [16][17][16,17]. However, the best way to use nanotechnology in agricultural applications has been limited by a lack of mechanistic understanding of how plants absorb, mobilize, and respond biologically to NPs. The type, particle size, composition, and specific surface area of these nanoparticles are the primary determinants of their effects on plants [18]. Plants’ reactions to NPs are exceptionally varied and subject to natural circumstances, plant species and age, duration and dose of exposure, and NP properties, such as structure, size, surface covering, morphology, stability, or disintegration, among different factors [19]. The integrated implications for specific NPs, including phytotoxic and beneficial effects on plants, have been precisely discussed in the literature [20][21][22][20,21,22]. Titanium oxide (TiO2) NPs, for example, lessen UV-B-induced oxidative stress [23]; they also stimulate the production of organic form of nitrogen from inorganic form, supporting nitrate assimilation and acquisition [24], and increasing photosynthesis rate [25]. In contrast, water acquisition and transpiration rate in corn plants is negatively impacted by TiO2 NPs [26]. In a similar manner, carbon nanotubes (CNTs) belong to a class of NPs that are capable of entering the seed coat and facilitating the acquisition of water by tomato plant seedlings, which is necessary for germination [27]. The synthesis of NPs, which exhibit fascinating effects on living systems and possess a variety of properties, has been attempted in a number of ways [28][29][30][28,29,30]. Likewise, many reports demonstrate the potential impacts of NPs on plants in terms of morphology and physiology [31], whereas there have been few endeavors examining the impacts of MNMs on plants at the genetic level. For example, soybean seedlings have been shown to be genotoxic to cerium oxide (CeO2) NPs, and onion seedlings have been shown to be genotoxic to silver (Ag) NPs; both of these nanoparticles have a gene-level effect on seedling growth [32]. In addition, the Oryza sativa seedling’s flowering time was extended when carbon (C) NPs were combined with natural organic matter [33]. Zinc oxide (ZnO) NPs, which harm the epidermal and cortical cells of Lolium perenne, are another example [34]. It is crucial to establish the interactive mechanism to study the relation between plants and their responses against nanoparticles, especially related nutrient flux, photosynthetic efficiency, and oxidative stress protections.
There are transcriptomics studies that assess the potential toxicity of nanomaterials in plants, and many of them focus on their impact on genes that respond to stress [35][36][35,36]. Because they show how NPs affect gene expression, the transcriptomic and proteomic studies add new information to our understanding of how plants interact with NPs. Carbon nanotubes (CNTs) were used in a study on tomato seedlings, and upregulation of various genes involved in stress-related processes was uncovered through microarray analysis [37].
A comparison examination of changes in total expression, however, points to a minimal effect on the transcriptome [38][39][38,39]. Biotic and abiotic stresses like pathogens, salinity, or drought may cause higher transcriptional reactions than nanoparticles [40][41][40,41]. It is obvious that plants did not have the opportunity to create unique gene regulation in response to novel nanomaterials; instead, they use regulatory circuits that are shared with other stress responses [42]. The suppression of genes for root development and pathogen response is a common consequence of abiotic stress [43]. There are reviews of further works that support these findings [44][45][44,45]. In a similar vein, additional research has shown that in Arabidopsis thaliana seedlings, gene expression for the peroxide-scavenging enzyme was influenced by CNTs [46]. Injection of single-walled carbon nanotubes (SWCNTs) to leaves increased transcription level for the two enzymes: ascorbate peroxidase (APX1) and manganese superoxide dismutase (Mn SOD1). Arabidopsis thaliana was the subject of a study using ZnO and TiO2 NPs, with the former exhibiting phytotoxicity in the aforementioned plant and the latter on depicting profoundly significant positive effect on the plant [24][34][47][48][49][24,34,47,48,49]. Researchers have utilized advanced techniques to find NP uptake in plant tissues upon root or foliar exposure [50][51][50,51]. Such examinations have really demonstrated that some NPs enter through roots or passes on and can cross natural hindrances to move inside plants, in contrast to their conventional analogues [52][53][52,53]. But the molecular drivers of the uptake and transport of NMs in plants remains inconclusive.
To fully utilize the potential of NPs for sustainable crop production, it is essential to have a comprehensive understanding of the biological networks at multiple levels [54]. In recent years, the examining of NP–plant interaction has developed from conventional, single endpoint measures to discovery-oriented, high-throughput system biology approaches, referred to as “omics”. Analytical methods and bioinformatics tools have increased in sensitivity and accuracy [55]. “Omics” refers to the undistinguished screening of biomolecules in an organism, specifically genes (genomics), mRNA (transcriptomics), and proteins (proteomics). With the rising need for mechanistic comprehension of complex agronomic qualities and crops’ reaction to NPs exposure, omics technologies have picked up speed in agriculture and nanotoxicity studies. This worldview helps in framing a hypothesis by checking the reaction of biomolecules upon perturbing biological cycles with NPs, followed by a combination of worldwide datasets onto pathways utilizing advanced bioinformatics [56][57][56,57]. In this manner, researchers are expected to provide more comprehensive knowledge to figure out the plant–NP interface.
2. Plant Interaction with Nanoparticles
Materials react substantially differently at the nanoscale than they do at larger dimensions, and they frequently exhibit unusual chemical and physical characteristics [41]. This makes the transformation of matter through nanotechnology a promising area for the creation of new goods and procedures. On the other hand, their distinct features enable innovative materials to interact with biological systems in unanticipated ways [42][43][42,43]. Different plants interact with NPs through chemical or physical contact (Figure 1). These interactions result in physical and chemical signaling, which generates new molecules, such as reactive oxygen species (ROS), which may pose serious damage to plants [58]. These ROS cause oxidative stress and damage [59], altering the movement of ions throughout the cell membrane [60], and peroxidation of fatty acids [61]. The key factors that determine plant–NP interaction are the nanoparticle’s characteristics, environmental factors, and plant species [62]. It is still unclear how the first NP contact point in plants chooses the traffic route. Nevertheless, a number of studies have suggested a few paths that could enhance theour comprehension of nanoparticle trafficking. The well-known channels that determine the entry route are direct application, contaminated sediments or soil, accidental release, and atmospheric fallouts. Xylem, cortex, and pericycle play crucial roles in facilitating nanoparticles’ entry into the plant via the root [63]. Particle entry at various key checkpoints are controlled by a few key components. The cell wall is the first checkpoint, and it functions as a structural sieve, allowing only NPs with compatible pore sizes to enter. Only NPs with pore sizes comparable to those of the cell wall were able to successfully travel outside the cell membrane through the cell wall [13]. Through the root system, tiny NPs can quickly enter plant cells, while larger NPs can only enter through larger openings such as stomata, stigma, and hydathodes. The cell rarely receives larger NPs; as a result, there is no change in cell metabolism [64]. Metal-based NPs change into reactive metal ions when they enter plant cells, react with various functional protein groups, and disrupt the biochemical activity. These NPs easily adsorb various inorganic ions and molecules from the soil or nutrient medium due to their large surface area, leading to indirect toxicity symptoms such as wilting and chlorosis [65]. The reaction between NPs and plants determines their accumulation and transfer across various trophic levels.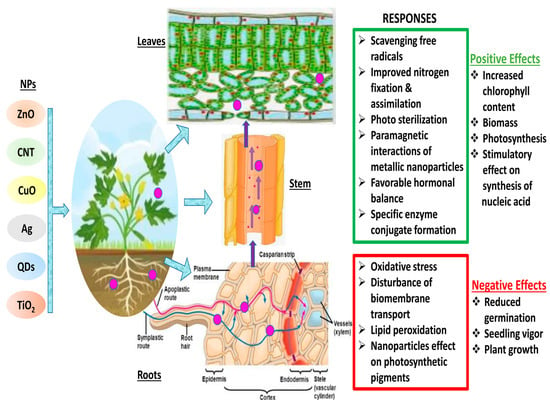
Figure 1. Schematic diagram of plant—NPs interactions and various responses. (Red color indicates the negative impacts of NPs interaction in plants, whereas green color shows the positive interactions of NPs in plant cells)
3. Nanoparticle–Plant Interaction: Transcriptomic and Proteomic Debate
3.1. Genomic Insight into Nanoparticle–Plant Interaction
Since nanotechnology has been recognized as the most cutting-edge, rapidly developing, and labor-saving technology by the scientific community as of late, it now has applications in almost every field of science. The toxicity caused by the constant deposition of NPs in the environment is not ignorable, especially in terms of plant performance. NPs interaction at genomic level is shown in Figure 2. A bulk of the literature demonstrates the morphological and physiological phytotoxic effects of NPs, but very little demonstrates the gene-level toxicity of NPs. Researchers reported that CuO NPs were toxic to some agricultural crops like Loliumrigidum, Loliumperenne and Raphanussativus, Lolium rigidum, Lolium perenne, and Raphanus sativus, and after prolonged NP exposure, DNA was destroyed. The deposition of compounds that have been altered by oxidation led to the formation of mutagenic DNA lesions, which in turn caused rapid disruptions in plant growth [66][121]. Similarly, ZnO NP toxicity in wheat seedlings was examined and it was discovered that nitric oxide (NO) mitigates this toxicity in wheat effectively [67][122]. Additionally, figuring out the NPs’ genotoxic endpoints is a concern because they have been linked to metal’s genotoxic effects in plants [68][123], but any generalizations regarding NP-induced phytogenotoxicity must be handled with care.
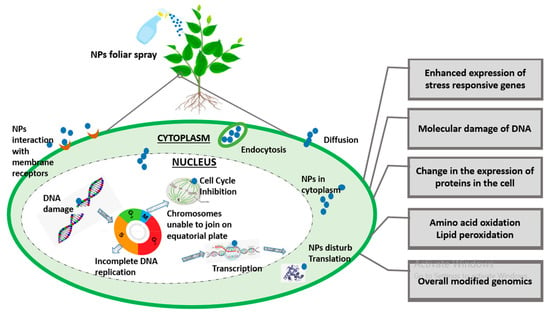

Figure 2. A brief illustration of genomic insight into plant–-NPs interaction.
3.2. Genes Regulation with Respect to NP-Induced Genotoxicity
At the molecular level, the basis of nanoparticle-induced phytotoxicity in higher plants is still poorly understood. However, transcriptomic analysis, upon NP exposure, has presented information about the up- and downregulation of different genes, which has provided some knowledge regarding NP toxicity at the genetic level in higher plants. For example, it was experimented that when maize root tissue was exposed to single-walled carbon nanotubes (SWCNTs), the genes for RTCS and SLR1 were upregulated and the genes for RTH3 and RTH1 were downregulated. Aquaporin also plays a significant role in nanoparticle acquisition. A variety of stress parameters control or regulate many stress-inducible genes in plants [69][124]. In another study, it was found that an increase in aquaporin production in tomato seedlings upregulated the water channel and NtLRX1 genes [70][125]. Some researchers treated Eruca sativa with Ag NPs, recorded the proteomic profile of Ag-treated plants, and different gene and protein expressions. Ag NPs effected the number of proteins in the plant cell’s vacuole and endoplasmic reticulum (ER), indicating that these organelles are Ag NPs’ primary targets [71][126]. When Arabidopsis plants were treated with varying concentrations of gold (Au) nanoparticles, similar expression results for micro RNAs (miRs) were observed [72][127]. When Au NP exposure was compared to a control, expression of some microRNAs, especially miR164, miR395, miR167, miR408, and miR398, were found to be downregulated. However, this study’s findings are in direct opposition to those of Burklew, Ashlock [73][128] regarding tomato plant seedlings in significant ways. This further proves that NPs have a species-specific phytotoxicity. Moreover, NPs alter gene expression even at very low concentrations, suggesting that the impact of NP-caused toxicity at cellular level can be gauged using gene expression analysis [74][75][129,130]. In this way, gene expression analysis assists in the identification and development of transgenic plants that are sensitive or tolerant to certain stresses [76][77][131,132]. As a result, genomics analysis is profoundly useful for determining the mechanism of NP toxicity and relating any physiological or morphological changes at the genetic level in plants. The fundamental processes of electron transport chain signaling are disrupted by the NP-induced phytotoxicity, which ultimately affects the organism’s cell cycle.
3.3. Transcriptomic Insight into Nanoparticle–Plant Interaction
Numerous researchers have reported evidence of nanoparticle-induced alterations in plant gene expression. It is averred that exposure to ZnO and TiO2 altered the gene expression found in Arabidopsis thaliana roots. They concluded that ZnO NPs are the most toxic NPs among the three abovementioned NPs because they induced downregulation and upregulation of 826 and 660 genes for stress regulation, respectively, although the least toxic TiO2 NPs only had an impact on 74 and 80 genes, respectively [78][118]. Despite the researchers’ clear demonstration of up- and downregulation of genes caused by the phytotoxicity of nanoparticles, comprehensive details have not yet been portrayed. In a recently conducted study, they exposed A. thaliana to various sizes and types of NPs under abiotic and biotic stresses to observe the early changes in gene expression. As a general response to nanoparticle-induced stress, they observed transcriptional repression and downregulation of gene expression [79][134]. In another study, examination of proteomic, ionomic, and transcriptomic shifts have put Arabidopsis into focus to determine how Au NPs affect growth and development. The expression of the MYB, BHLH, and WRKY gene families, which are involved in homeostasis of some essential metals and Fe, differed according to the transcriptome analysis of Arabidopsis roots [80][153]. It was inferred that Au impacted the normal homeostasis of essential elements like Mn, Fe, and Zn. By observing genetic and physiological responses of plants to Au, it has also been discovered that Au is uptaken by Arabidopsis and then transported to the shoot. In addition, it has been observed to have an effect on the transcription of a specific group of genes, particularly cation transporters, with 869 genes being upregulated and 851 genes being downregulated. IRT1 and IRT2, iron-regulated transporters, have shown the lowest expression of genes. Downregulation of eleven aquaporin genes was also reported in this study. Metal uptake, particularly that of heavy metals, is thought to involve a number of these genes [81][154]. Numerous studies have looked into the different kinetics that NMs use to acquire, translocate, and internalize NMs, as well as the cytotoxicity that is recorded at the level of gene expression. These studies also found that NMs affected plant growth and development, along with a number of other morphological and physiological effects. Although the genetic processes underlying these effects have not yet been discovered, it has been observed that these effects are species-specific. The transcripts of genes involved in vacuolar proton or cation exchange, hormonal responses, antioxidant systems, pathogen-responsive genes, decreases in photosynthesis genes’ transcript formation, aquaporin genes, as well as biogenesis and cell organization genes, were found to be upregulated in NP-mediated toxicity.
3.4. Proteomic Insight into NP–Plant Interaction
Over the past few decades, various multidisciplinary studies on the impacts of Ag NPs on plants have shown that they are toxic at many levels—cellular, morphological, physiological, etc. However, only a small number of studies have examined the detrimental proteomic effects of Ag NP stress on plants. In an effort to carry away the toxicity posed by Ag NPs, a gel-based proteomic study of Oryza sativa seedlings was recently conducted [82][157]. Ag NP-receptive proteins of were reported to be primarily associated with Ca2+ signaling and regulation, the oxidative stress response pathway, transcription, protein degradation, cell wall synthesis, cell division, and apoptosis. Specific defense-related proteins, i.e., SOD, glutathione S-transferase (GST), and L-ascorbate peroxidase, were found to be overexpressed after exposure to Ag NP toxicity. Resultantly, this accelerated the generation of reactive oxygen species (ROS) under Ag NP treatment. Previous research has suggested that secreted ions or Ag NPs could disturb the cell’s metabolism by binding to Ca2+/Na+ATPases and Ca2+ ion channels—the secondary messenger receptors. In a similar manner, a proteomic analysis of Eruca sativa roots exposed to AgNO3 and Ag NP stress revealed that both bulk metal and NPs can alter the structure of proteins associated with redox regulation and ultimately disrupt cellular homeostasis [71][126]. However, the fact that the exposure of Ag NPs is sufficient to alter the membrane proteins of vacuoles and endoplasmic reticulum suggests that these NPs primarily target both of these specific organelles. According to these findings, Ag NP phytotoxicity is caused by its unique chemical and physical properties rather than its continuous release to the atmosphere [71][126]. The agricultural sector is an important field in terms of the application of gold nanoparticles. After exposing Arabidopsis thaliana to Au NPs, a proteome analysis revealed the induction of glutathione S-transferases in the shoot. Additionally, researchers have also suggested that auxin, the plant hormone, plays a significant role in shaping Au-induced root system architecture [80][153]. In a similar way, Ag NPs at three distinct particle sizes (2, 15, and 50–80 nm) and concentrations (0.2, 2, and 20 ppm) were used in a series of experiments on soybean seedlings [83][158]. They demonstrated that, under flood stress, Ag NPs of 15 nm particle size promoted soybean seedling growth rate and germination potential in comparison to larger and smaller Ag NPs. The proteins altered by the exposure of Ag NPs were mostly linked to stress signaling, the detoxification pathway, and cellular metabolism. The concentration of these proteins increased under flood stress, but the proteins rapidly decreased when exposed to Ag NPs. Additionally, treatment with Ag NPs demonstrated a shift in metabolism from fermentative to cellular processes. According to the findings, soybean seedlings subjected to Ag NP treatment under flooding stress experience less oxygen loss as a result of treatment with Ag NPs, which have a particle size of 15 nm and 2 ppm. Additionally, soybean seedlings were found to be damaged at elevated Ag NP concentrations with particle sizes of 15 nm [83][158]. A comparative study with Ag, Al2O3, and ZnO NPs on two-day-old soybean seedlings subjected to flooding stress was recently conducted [84][159]. Surprisingly, treatment with 50 ppm Al2O3 NPs resulted in increased vigor and growth in soybeans. The primary functions of these Al2O3NP-responsive proteins were lipid metabolism, glycolysis, protein degradation, and protein synthesis. Under treatment with Al2O3 NPs, a fivefold increase in the negative NmrA-like transcriptional regulatory protein family was also observed. The proteomic results suggest that the growth of flood-stressed soybean seedlings may be attributed to regulation of energy metabolism and reduced proliferation of root cells. Similar to Ag and Au NPs, various cell systems have been used to investigate other NP toxicities. For instance, it has been demonstrated that CeO2 NPs inhibit maize growth by hampering stress-responsive protein expressions like CAT, heat shock protein, and APX [85][84]. Mediation of NP toxicity has been reported by the decline and destabilization in cell membrane potential, lowering ATP levels in cells, elevating stress-responsive and quorum-sensing proteins, expression of oxidative stress-tolerant proteins, Ca2+ release and its signaling, transcriptional proteins, as well as downregulating essential proteins related to growth-regulating mechanisms like cell metabolism, photosystem-II functioning, and sulfur assimilation. A brief description of NPs interaction at transcriptomic and proteomic level is shown in Figure 3.
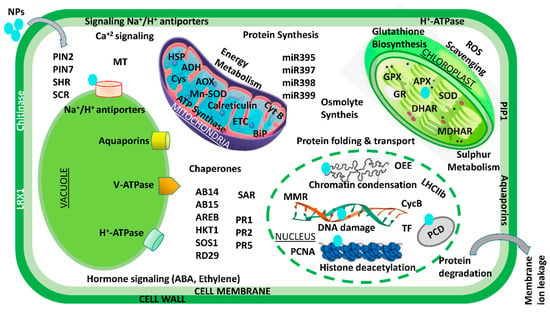

Figure 3. An illustration of transcriptomic and proteomic analysis of the plant–-NPs interaction. ABI: ABA-insensitive; AOX: alternate oxidase; ADH: alcohol dehydrogenase; AREB: ABA-responsive element-binding proteins; APX: ascorbate peroxidase; ARGOS: auxin-regulated gene involved in organ size; CycB: cyclin B; BiP: binding immunoglobulin proteins; Cys: cysteine synthase; GR: glutathione reductase; ETC: electron transport chain; GlyII: glyoxalase II; HKT1: high-affinity K+ transporter 1; GPX: glutathione peroxidase; HSP: heat shock protein; miR: microRNA; MDHAR: monodehydroascorbate reductase; MMR: mismatch repair gene; OEE2: oxygen-evolving complex 2; MT: metallothionein; PCNA: proliferating cell nuclear antigen; PIN: peptidyl–prolyl cis–trans isomerase NIMA-interacting; PCS1: phytochelatin synthase 1; PIP1: plasma membrane intrinsic protein 1; RbcL: large subunit of Rubisco; PR: pathogenesis-related; RD29A: responsive to desiccation 29A; SAR: systemic acquired resistance; ROS: reactive oxygen species; SCR: scarecrow; SOS1: salt overly sensitive 1; TF: transcription factors; SOD: superoxide dismutase; SHR: short root.
