Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Antonios Georgas and Version 3 by Jason Zhu.
The COVID-19 pandemic highlighted the importance of widespread testing for SARS-CoV-2, leading to the development of various new testing methods. However, traditional invasive sampling methods can be uncomfortable and even painful, creating barriers to testing accessibility.
- COVID-19
- sensors
- biosensors
- machine learning
1. Machine Learning-Enhanced Biosensors
Machine learning-enhanced biosensors represent a promising approach to improve the sensitivity and specificity of non-invasive SARS-CoV-2 testing. Biosensors are analytical devices that can detect and quantify biological molecules, such as viral RNA or antigens, in a sample. However, several factors, including sample quality, background noise, and other sources of interference, may limit the accuracy and sensitivity of biosensors. As shown in Figure 1, machine learning algorithms can be integrated with biosensors to enhance their performance by analyzing large amounts of data and identifying patterns that can be used to improve the accuracy and sensitivity of the biosensor [1][2][15,34].
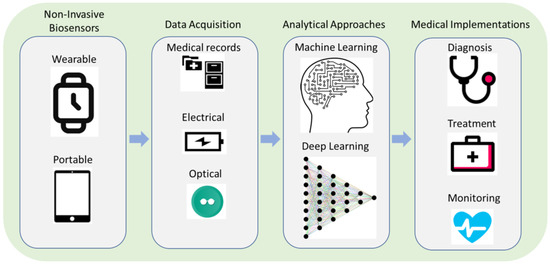
Figure 1. Non-invasive biosensors are enhanced with ML algorithms for medical applications.
2. Pattern Recognition and Error Detection
To overcome the limitations of biosensors in detecting low concentrations of SARS-CoV-2 in saliva samples, machine learning algorithms can be used to perform automated quality control of biosensor measurements. By analyzing large datasets, machine learning algorithms can identify patterns in the signal that correspond to the presence of the target analyte (in this case, SARS-CoV-2). Once these patterns are identified, the biosensor can use them to detect the presence of the virus more accurately in saliva samples. Employing supervised learning algorithms, such as support vector machines (SVMs) [3][35] (Figure 2a) or random forests [4][36] (Figure 2b), is one way to make use of these patterns. These algorithms can classify each measurement as either accurate or erroneous after being trained on a sizable dataset of biosensor readings with known ground truth values. The algorithm learns the fundamental patterns and relationships in the data throughout this training process, which enables it to predict new measurements with accuracy. The accuracy of the test results can be increased by highlighting measurements that the algorithm finds to be inaccurate. This strategy has the potential to completely transform the field of biosensing and showed considerable promise in terms of enhancing the precision and dependability of biosensors, notably in the context of non-invasive SARS-CoV-2 testing. Specific examples will be given in SectionSection 4 4 of using supervised algorithms such as SVMs to improve SARS-CoV-2 biosensors by recognizing specific patterns in a dataset and assigning the data to the correct class [5][6][37,38].
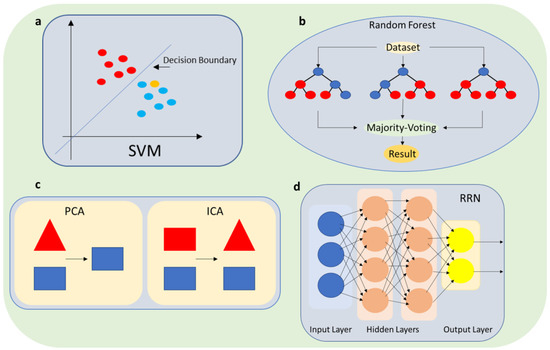
Figure 2. Schematic illustration of ML algorithms; (a) SVM; (b) random forest; (c) PCA-ICA; and (d) RRN.
Moreover, machine learning can also enable biosensors to detect errors or noise in the measurement process. Noise in the data can arise from various sources, such as variations in the sample matrix or environmental factors such as temperature and humidity. By analyzing patterns in the data and comparing them to known signal patterns, machine learning algorithms can distinguish signal from noise and reduce false positives and false negatives in the measurements, as shown in Figure 3. Specifically, unsupervised learning algorithms such as principal component analysis (PCA) [7][8][39,40] or independent component analysis (ICA) [9][41] (Figure 2c) can be used to identify patterns and correlations in the biosensor measurements and separate them from noise. Experimental results [10][11][42,43] show that by using these algorithms, the biosensor can better discriminate between the signal from the virus and other noise sources, such as background fluorescence in the case of sensitive detection of porcine epidemic diarrhea virus [10][42], or even facilitate the classification of viruses based solely on their intrinsic spectra [11][43], as well as improve the sensitivity and specificity of the test. PCA was also used successfully to enhance the biomarker selection process and improve the performance of the classifier in distinguishing between different classes of proteins and viruses [12][44].
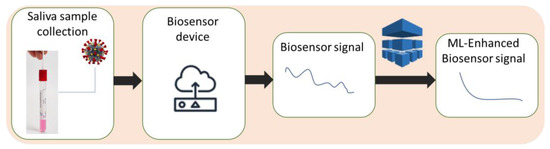
Figure 3. Machine learning algorithms improve biosensor signal by distinguishing it from measurement noise.
Thus, machine learning-enhanced biosensors can provide a more accurate and reliable non-invasive method for detecting SARS-CoV-2 in saliva samples. The ability to detect low concentrations of the virus with high specificity and sensitivity could greatly improve the accessibility and effectiveness of testing efforts, particularly in areas where invasive testing methods are not readily available or feasible.
3. Contrastive Learning
Contrastive learning is a powerful technique used in machine learning to help models learn to distinguish between different types of data points [13][14]. The main goal of contrastive learning is to create pairs of data points that are similar in some way (positive pairs) and pairs that are dissimilar (negative pairs). By training models to distinguish between these two types of pairs, researchers can promote the closeness of similar pair representations and increase the orthogonality of dissimilar pair representations [14][45]. Contrastive learning can be applied to biosensors to learn data representations that highlight the distinctions between various signal kinds [15][46]. This is advantageous for biosensors since the signals they pick up are frequently rather weak and are easily masked by background noise or other forms of interference. Contrastive learning can help to increase the accuracy of biosensors by making it simpler to discern between various types of signals by teaching representations that highlight the contrasts between signals.
Contrastive learning can be a useful strategy for learning meaningful representations that highlight the differences between various signal types in the setting of biosensors, such as cardiac signals [16][47], electroencephalogram (EEG), and electrocardiogram (ECG) [17][18][48,49]. Weak signals that are easily obscured by background noise or other types of interference are common characteristics of biosignals. By using contrastive learning, the models can be trained to distinguish between various signal types based on their intrinsic contrasts, improving the accuracy of biosensors.
The unique difficulties posed in this field must be carefully taken into account when using contrastive learning for biosensors. Biosignal datasets frequently feature a small number of subjects—generally under 100—and noisy labeling. Cheng et al. suggested that a self-supervised approach based on contrastive learning can be modified to overcome these difficulties [18][49]. This strategy seeks to simulate biosignals with a smaller number of patients and less reliance on labeled data.
Intersubject variability can have a detrimental effect on model performance in the regime of restricted labels and individuals. Subject-aware learning methods can be used to lessen this problem. One method is to use a subject-specific contrastive loss, where the contrastive loss is calculated using distributions specific to the person. This motivates the model to discover representations that accurately reflect the distinctive features of each subject’s biosignals.
Additionally, subject invariance can be encouraged during the self-supervised learning process by using adversarial training. The model’s ability to generalize across diverse subjects and perform better as a whole can be improved by training it to be invariant to subject-specific variances.
Figure 4 depicts a comprehensive pipeline for applying unsupervised/contrastive learning in the context of biosensors.
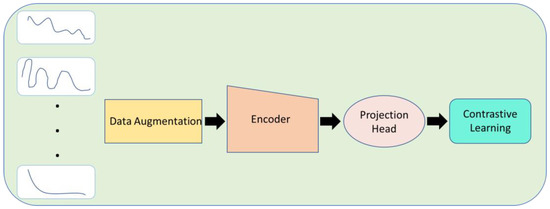
Figure 4. Contrastive learning highlights distinctions between various signals.
Data augmentation is the first process in the pipeline, starting with the biosensor signals as the input layer. To increase the diversity and robustness of the training dataset, this method entails producing extra varieties of the input data. Techniques for enhancing data may involve random translations, rotations, noise addition, or additional modifications.
The signals are then passed to an encoder after data augmentation. The encoder, which learns to extract meaningful and condensed representations from the input data, is a crucial component. It captures crucial features and patterns by transforming the high-dimensional signals into a lower-dimensional latent space.
The encoded representations are then supplied onto a projection head after the encoder. The separability and discriminative capabilities of the encoded representations are improved by the projection head’s additional mapping of the latent space to a separate feature space. The projection head may include one or more layers that subject the latent features to different non-linear transformations.
The pipeline culminates in contrastive learning, a self-supervised learning method that tries to reduce the agreement between dissimilar cases while increasing the agreement between comparable instances. The model learns to combine the encoded representations of similar biosensor signals and push apart the representations of dissimilar signals by utilizing contrastive learning. Through this procedure, the model is urged to acquire accurate representations of the underlying structure and properties of the biosensor data.
4. Real-Time Interpretation of Biosensor Measurements with Machine Learning
With the integration of machine learning (ML) techniques, biosensors can provide real-time interpretation of test results even after a few initial measurements, therefore providing a SARS-CoV-2 result in few seconds.
ML algorithms can be effectively trained to discern the specific signature of SARS-CoV-2 in saliva samples, effectively separating it from background noise and other interferences. Signal processing techniques play a crucial role in this process, with mathematical models such as recurrent neural networks (RNNs) (Figure 2d) being employed to extract meaningful information from biosensor signals [19][50]. Among these techniques, wavelet transform stands out as a valuable approach, enabling the decomposition of the biosensor signal into different frequencies and aiding in the discrimination between signal and noise [20][51]. RNNs excel in biosensor signal processing tasks due to their ability to capture intricate temporal dependencies within the data. The decomposition of the biosensor signal into distinct frequency bands, a common strategy in biosensing, enhances the differentiation between signal and noise. Leveraging the power of wavelet transform and other signal processing methods, machine learning algorithms can successfully extract valuable insights from biosensor signals, leading to more accurate and sensitive detection of SARS-CoV-2 in saliva samples.
Another proposed method that might significantly boost the effectiveness and precision of biosensor testing is adaptive sampling [21][22][52,53]. Based on the parameters of the saliva sample being examined, this method entails varying the sampling rate and length. For instance, the algorithm may lower the sample frequency and length if the biosensor identifies a strong signal from the early measurements because it is likely that the signal will stay high throughout the test. On the other hand, if the first measurements reveal a weak or noisy signal, the algorithm may raise the sample frequency and length to gather more data and improve the accuracy of the outcome [23][24][54,55]. Compared to conventional testing techniques, which generally include a predetermined sample rate and length, adaptive sampling provides several advantages. Adaptive sampling can decrease the time needed for accurate detection while simultaneously boosting the dependability of the result by modifying the sampling rate and duration based on the features of the sample. This is especially vital for applications that need quick and precise detection, including clinical diagnostics or environmental monitoring. By examining the characteristics of the saliva sample and modifying the sampling rate and length appropriately, machine learning algorithms may be trained to accomplish adaptive sampling. A sizable dataset of biosensor readings with well-known ground truth values is necessary for this approach in order to train the algorithm and enhance its performance. Once it is trained, the algorithm may be connected with the biosensor to deliver real-time adaptive sampling and boost testing precision and effectiveness.
