Regarding dinoflagellates, the mechanisms related to the bioluminescent phenomenon have been well studied; however, at present, some points that remain unknown still persist. That is the case of luciferin biosynthesis, which has been reported to be an intricate process with several metabolic pathways involved. In the context of this controversial scenario, various hypotheses about the biosynthesis of luciferin in dinoflagellates are presented.
- dinoflagellate
- bioluminescence
- luciferin
- P630
- blue compound
- glutathione S-transferase
Carlos Fajardo 1* and Marcos De Donato 2
1 Microbiology Laboratory, Institute of Viticulture and Agri-food Research (IVAGRO), Environmental and
Marine Sciences Faculty. University of Cadiz (UCA), 11510 Puerto Real, Spain; carfaqui07@yahoo.es
2 Tecnologico de Monterrey, Escuela de Ingenieria y Ciencias, 76130 Queretaro, Mexico; mdedonate@tec.mx
- Introduction
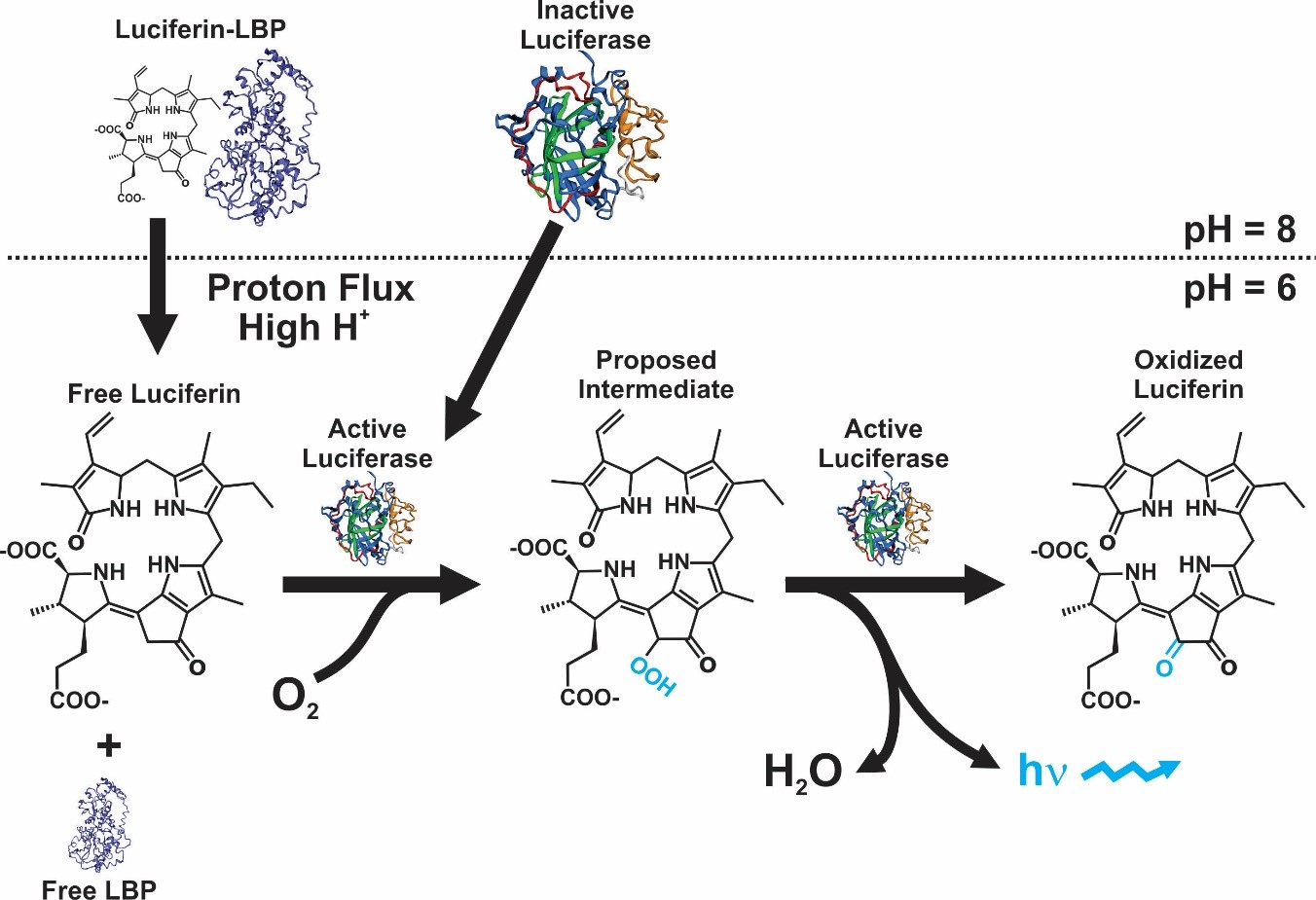
Although in a strict sense the mechanism is not equal in all bioluminescent organisms, the majority of them present the same basic chemical reaction. In this, the enzyme luciferase reacts with luciferin in the presence of O2, originating an oxyluciferin that release photons while decaying from a high to low energy state [1]. The genes and cellular mechanisms related with bioluminescence in dinoflagellates are unique and has been well-studied. The bioluminescent event occurs inside special organelles called scintillons, which contain the luciferase enzyme, the substrate luciferin, and in most of species a luciferin-binding protein. The role of this protein is to protect luciferin from oxidation by binding to it at physiological pH [2][3][4][5][6] (Figure 1).
An important fact that remains an enigma today concerns about the process related to the luciferin biosynthesis mechanism. In the case of Pyrocystis lunula, unlike other bioluminescent dinoflagellates, the levels of luciferin and luciferase are constant during the all circadian cycle [4], thus, in this species, the rhythm is related with modifications of their intracellular localization and reutilization, instead of daily de novo biosynthesis of all the components. The process of daily de novo biosynthesis and destruction mechanism has been reported in the case of other species such as Lingulodinium polyedra [7][8]. Based on the current evidence, it has been hypothesized that luciferin can be biosynthesized by different ways, and it is considered to be ¨universal¨ in dinoflagellates, since luciferin from any bioluminescent species of dinoflagellates can be used as a substrate to produce light [9]. Dinoflagellate luciferin was suggested to be a photo-oxidation degradation product of chlorophyll a [10]; nevertheless, this would not be the case in all species. For example, L. polyedra produce luciferin only during the dark period, that is to say when photo-oxidation is impossible [6]. Moreover, the heterotrophic species Protoperidinium crassipes can retain its bioluminescence for up to one year without foods carrying luciferin or chlorophyll [11], and thus, it can be pointed out that it produces luciferin generated from another precursor. In view of this scenario, it is clear that there is more than one mechanism responsible for luciferin production [6][9].
Figure 1. Bioluminescence reaction in dinoflagellates [9].
- Luciferin
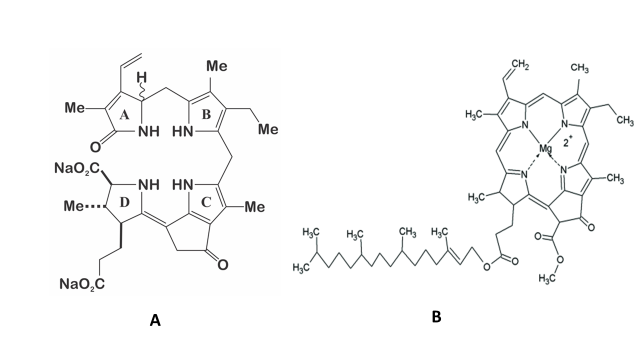
The structure of the molecule of dinoflagellate luciferin is a tetrapyrrole-type (Figure 2), comparable to chlorophyll a, and krill luciferin (Euphasia superba) [12][13]. This molecule is highly labile to oxidation (included photo-oxidation), low pH, and at high salt concentration [14]. It has been reported that P. lunula produce more luciferin than any other bioluminescent dinoflagellate [13], even one hundred times more than L. polyedra [4][13], which is why it has been established as a model organism for the study of this molecule. Moreover, luciferin from P. lunula can cross-react with the luciferases of several species of bioluminescent dinoflagellates. Interestingly, it can actually cross-react with the bioluminescent system of euphausiid shrimp E. superba [2][15][10].
Figure 2. A) Structure of the dinoflagellate luciferin; B) Structure of the chlorophyll a.
Regarding P. lunula, it was hypothesized that luciferin is a photo-oxidation breakdown product of the chlorophyll a [10]. Based on this hypothesis, Liu and Hastings [16] proposed that dinoflagellate heterotrophic species can take the luciferin through food, straightly or by degradation of chlorophyll of the preys. Additionally, Buskey et al. [17] carry out studies on heterotrophic dinoflagellates to define the effects of light (photoinhibition) and starvation on their bioluminescence. The bioluminescence of Noctiluca scintillans was not restricted by exposure to light, nor was there any detectable diel pattern of stimulable bioluminescence. N. scintillans fed with a mixture of phytoplankton foods, slightly enhanced in bioluminescence capacity over a two weeks’ period, whereas cells that were kept without food exhibit a continual decrease in bioluminescence. Furthermore, it was found that all bioluminescent species of Protoperidinium have their mechanically stimulable bioluminescence inhibited by the exposure to light. The recovery of bioluminescence of P. covergens was complete within thirty minutes of transfer to dark and complete photo-inhibition take place ten minutes after exposition to light. Apparently, there’s not a mechanically stimulable diel pattern of bioluminescence in P. divergens. On the other hand, P. depressum maintained for three days without food, display a decrease in the bioluminescence capacity; however, the same species maintained under different food concentrations for 48 h did not manifest variance in its bioluminescent potential [17]. Therefore, the suggestion that luciferin is produced from photo-oxidized chlorophyll [10] would only be true for some species, like P. lunula, which keeps its luciferin during all the daily cycle. In contrast, L. polyedra only produces fluorescent luciferin during the beginning of the night [5], so, its biosynthesis cannot be explicated by a photo-oxidation process. Additionally, the heterotrophic species P. crassipes can preserve its bioluminescence for an extremely long period of time, even one year, without chlorophyll or luciferin containing food [11] and, thus, must biosynthesized luciferin using another precursor. Moreover, Wu et al. [18] have confirmed the intracellular production of luciferin in P. lunula using amino acid tracers. Thus, it is probably that more than one mechanism is related to the biosynthesis of luciferin [6][9][19][20].
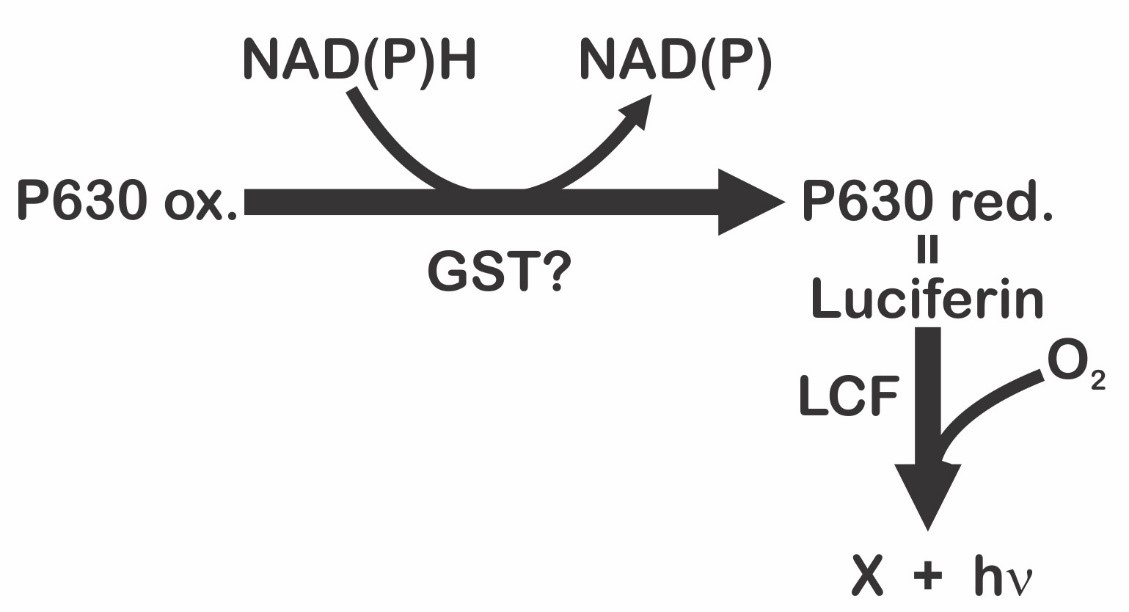
On the other hand, Fresneau and Arrio [20] maintain that bioluminescence in P. lunula is ruled by the reduction state of the luciferin precursor. The precursor of luciferin in this case would be the compound P630 (named in this way by his maximum excitation wavelength: 630 nm). Moreover, it was demonstrated that P630 and luciferin have the same peptide moiety. P630 is a chromo-peptide more stable than luciferin. P630 is made up by a polypeptide of 4.8 kDa, and a linear tetrapyrrole (600 Da). Cations could oxidize P630 or cleave the bond between the peptide chain and the extended tetrapyrrole. Reduction of P630 could be achieved enzymatically by a NAD(P)H-dependent oxidoreductase, or chemically by 2-mercaptoethanol or dithiothreitol. The state of reduction lead to a pH-dependent conformational change in the chromo-peptide. Furthermore, reduced P630 shows the same spectral features as the luciferin. Additionally, luciferase is able to oxidize the reduced P630 producing light emission. Interestingly, luciferin (at -20 °C on methanol), is partly and spontaneously transform into P630. These observations suggest that interconversion P630-luciferin could represent the oxide-reduction equilibrium [20]. These data pointed out that reduced P630 is a luciferin, and, therefore, the precursor of luciferin will be the P630 oxidized form [21]. According to these authors [20], the bioluminescent event is the result of an intricate mechanism ruled by two sequential reactions (Figure 3). The first is represented by the reduction of the luciferin precursor P630 mediated by a NAD(P)H-dependent reductase [21]; being the second the classical and previous described luciferase-luciferin reaction (Figure 1). As P630 could be reversibly reduced, it seems to be an important interchange point of reducing power that imply a novel electron transfer pathway. According to Fresneau and Arrio [20], the electron transfer mechanism that regulates the oxide-reduction balance of P630 should be considered as the source of luminescence. Moreover, it has been reported that P630 seems to be related with others intricate light-modulated reactions [22].
Figure 3. Light emission process in dinoflagellates proposed by Fresneau et al. [21], and modified by Fajardo et al. [9].
Interestingly, Nakamura et al. [12] also noted the presence of a peculiar product with a deep blue color during the isolation and purification process of dinoflagellate luciferin. This substance, called in this case blue compound, was isolated and purified. In contrast with the luciferin, Nakamura et al. [12] points out that blue air-oxidation product revealed its UV-visible absorption maxima at 633 and 590 (shoulder) nm, indicating the existence of a chromophore more conjugated. The UV-visible spectrum reported in this case for the blue compound was the following: (80% methanol containing 0.1% NH40Ac) 234, 254, 315, 370, 410, 590 (shoulder), and 633 nm; and the FAB mass spectrum was (glycerol) m/z 587 [(M - 2Na + 3H) +], 609 [(M - Na + H)’], and 631 [(M + H)’]. These data are in line with the observations made by Fresneau et al. [21] in relationship with P630: maxima absorption of P630 (oxidized) 630-370-315-250 nm; P630 (reduced) 390 nm; P630 maximum fluorescence excitation 630 nm. According with this information, Fajardo et al. [9] suggested that precursor P630 or blue compound, and luciferin are different stages of the same molecule.
According with Fresneau and Arrio [20], dinoflagellate bioluminescence could be considered as a metabolic process related with the control of excess intracellular reducing power generated by photosynthesis and respiration. This hypothesis is in line with the discovery of a specific chloro-respiration system reported by Bennoun [22], and with other reports made in cyanobacteria [23] of an alternative respiration system linked to photosynthetic thylakoid membranes [24].
Recently, Wang and Liu [13] proposed an alternative mechanism of dinoflagellate luciferase catalysis linked to a Dexter energy transfer system. These author hypothesized that an excited state oxyluciferin intermediate produced by the luciferase catalysis could transfer energy to another molecule of luciferin, or an analog, which, by relaxing with the radiative emission of light, would then serve as the bioluminophore. However, Ngo and Mansoorabadi [25] suggested that an excited state intermediate produced by the reaction between luciferin and oxygen, an excited state gem-diol(ate) intermediate, can serve directly as luminophore. These authors point out that a gem-diol(ate) intermediate as the bioluminophore, and also suggest that if the luciferase catalytic process starts with the more stable E-isomer of luciferin, the mechanism presumably involve a Chemically Initiated Electron-Exchange Luminescence (CIEEL) system. As in the case of fireflies, this system has been mention to explain other bioluminescent reactions [26][27][28]. On the other hand, if luciferin has the Z-configuration, these authors suggest that a twisted excited state gem-diol(ate) intermediate could serve as the bioluminophore, and if this were the case, the luciferase would catalyze a Twisted Intramolecular Charge Transfer (TICT) reaction. TICT states have been linked to diverse photochemical reactions but until recently had not been related to any bioluminescent system [25][29]. According with Fajardo et al. [9], the blue compound-P630 molecule could be related with this gem-diol(ate) intermediate. However, this is a hypothesis that needs more investigation.
On the other hand, as stated above, dinoflagellate luciferin is structurally very similar to chlorophyll a [10], and in fact the relationship between their biosynthesis has been demonstrated [18]. Janouskovec et al. [30] suggest that, from a metabolic point of view, all free-living dinoflagellates are dependent on plastids. Therefore, plastid tetrapyrrole biosynthesis would explain the existence of luciferin in non-pigmented dinoflagellates. Janouskovec et al. [30] also suggest that a single tetrapyrrole pathway, of a predominantly plastid origin that begins from glutamate, is present in all core dinoflagellates. This is a typical characteristic of eukaryotic plastids [31]. Non-photosynthetic species such Noctiluca, Dinophysis, and Oxyrrhis, also present membrane translocators genes for triose phosphate, ferredoxin redox, and plastid iron–sulfur systems [30]. The conserved presence of signal peptides and N-terminal extensions are related to a plastid origin. According to these authors, the plastid tetrapyrrole pathway is indispensable for heme biosynthesis in all core dinoflagellates, and thus, independently of the presence of photosynthesis, could be related to the luciferin production in any bioluminescent dinoflagellate. This would explain the biosynthesis of luciferin produced from an earlier intermediate of the chlorophyll biosynthesis pathway, from a chlorine-like tetrapyrrole or chlorophyllide. These finding reinforces the hypothesis that non-photosynthetic bioluminescent dinoflagellates rely on a biosynthetic pathway derivative from heme and chlorophyll production [30].
According to Martin et al. [32], the first ecosystems on earth, fueled by geological H2 for CO2 fixation, were chemotrophic. Those demanded flavin-based electron bifurcations to reduce ferredoxin, and probably the ancestral photochemically active pigments were Zn-tetrapyrroles. These authors hypothesized that after the transition of red-absorbing chlorophyll-like pigments, the original system of action was linked to a light-driven electron transport chain that reduced ferredoxin by a reaction center progenitor through H2S and electrons. Martin et al. [32] also suggested that photosynthesis eventually emerge in an ancestral anoxygenic cyanobacterial progenitor, being the chlorophyll a, the original configuration [32]. Furthermore, the biosynthesis of chlorophyll a and heme, as well as the bilin pigments produced from it, presents common steps that require oxygen for catalysis, and need to be carried out by the oxygen-dependent coproporphyrinogen III oxidase [33].
- Glutathione S-Transferase
In P. lunula, the antioxidant enzyme glutathione S-transferase (GenBank AAN85429.1) [34] shows a pfam05295 domain (Luciferase_N), which also present a high level of conservation with the sequences reported in the case of luciferase and luciferin-binding protein. Moreover, it was hypothesized the exon recombination as a possible explanation for this homology [4]. Nevertheless, the function of this N-terminal domain on P. lunula glutathione S-transferase remains undetermined [34]. Additionally, a glutathione S-transferase -N-Sigma-like domain, belonging to the thioredoxin-like superfamily, is also reported. Functioning as protein disulfide oxidoreductases, the members of this group can modify the redox state of target proteins by the reversible oxidation of their dithiol active site. The thiol of the cysteine, in the reduced state, can donate a reduction equivalent to other unstable molecules [34] and highly reactive oxygen species like luciferin [9].
Glutathione S-transferases include a vast family of eukaryotic and prokaryotic isozymes that have the capacity to catalyze the conjugation of the reduced form of glutathione to xenobiotic substrates for detoxification [35][36][37]. According to Fajardo et al. [9], the glutathione S-transferase would be the NAD(P)H-dependent reductase that rules the state of reduction of P630 (Figure 3). The domain pfam05295 (Luciferase_N) is a common feature between glutathione S-transferase, luciferin-binding protein, and luciferase in P. lunula; nevertheless, the glutathione S-transferase sequence has not been reported in other Gonyaulacales. Studies centered on the NADPH-dependent detoxification point out that eukaryotic cells use diverse mechanisms to manage with the negative effects produced by reactive carbonyls. The glutathione S-transferases pathways, which are involved with the glutathione or thioredoxin redox cycle, conjugates aldehydes with glutathione, performing a key role for detoxification [38]. Some of the glutathione S-transferase proteins are localized in the mitochondria and chloroplasts; and because chloroplasts contain large quantities of glutathione, the glutathione-dependent detoxification system is considered highly efficient [39]. Nevertheless, the presence of the domain pfam05295 (Luciferase_N) in the glutathione S-transferase has only been reported in the case of P. lunula [34][40]. If indeed this protein species is implicated in the mechanism of controlling the oxidative degradation of luciferin, this could explain the fact that P. lunula contains more luciferin than any other bioluminescent dinoflagellate [4].
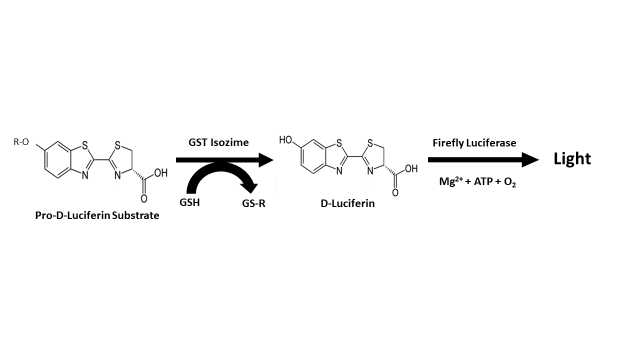
At this point, it is very important to note that in the case of the optimization and miniaturization of a high-throughput homogeneous method using glutathione S-transferases from three different species (human isozymes A1-1, M1-1, and P1-1, mouse isozyme A4-4, and the major glutathione S-transferase from the parasitic worm Schistosoma japonicum); Yasgar et al. [41] have reported a very similar action system (Figure 4) to the one proposed by Fajardo et al. [9], in relation to the possible role of glutathione S-transferase in the process of biosynthesis of luciferin in dinoflagellates (Figure 3). This method is based on the use of pro-D-luciferin substrate from firefly for the measurement of glutathione S-transferase activity, configuring a sensitive screening assay via coupled luciferase reaction and standard luminescence detection [42][43]. Unlike the bioluminescent system of dinoflagellates, the system used by fireflies (Photinus pyralis, Luciola italica) requires ATP and Mg2+ as a cofactors of the enzymatic reaction that leads to the luminescent event; a fact that has led to the development of various biotechnological applications that take advantage over the peculiarities of the bioluminescent system of this group of organisms [44][45][46][47][48][49][50][51][52][53].
Figure 4. Assay principle reported by Yasgar et al. [41] for the measurement of glutathione S-transferase (GST) activity.
- Conclusions
The review of the available information suggests that is possible than more than one mechanism is related with the biosynthesis of luciferin of dinoflagellate. The link between the biosynthesis of chlorophyll and luciferin has been demonstrated; and apparently, plastid tetrapyrrole pathway is indispensable in all core dinoflagellates, therefore could be related to luciferin production in any bioluminescent dinoflagellate, independently of the presence of photosynthesis. For the model organism used in studies with the dinoflagellate luciferin, the species P. lunula, P630 or blue compound is one of the precursors of luciferin, and, at least in this species, the glutathione S-transferase protein could be involved in the biosynthesis process of this molecule. Various electron transfer mechanisms have been linked with the biosynthesis of dinoflagellate luciferin. These include the Dexter energy transfer, and depending on the luciferin configuration (E or Z), a CIEEL or TICT system, respectively.
Author Contributions: Conceptualization, C.F.; investigation, C.F. and M.D.D.; data curation, C.F.; writing—original draft preparation, C.F.; writing—review and editing, C.F. and M.D.D.; visualization, C.F. and M.D.D. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest: The authors declare no conflict of interest.
Entry Link: https://www.mdpi.com/1422-0067/21/5/1784/htm
Abbreviations
CIEEL Chemically Initiated Electron-Exchange Luminescence
GSH Glutathione
GS-R Glutathione Reductase
GST Glutathione S-Transferase
TD-DFT Time-Dependent Density Functional Theory
TICT Twisted Intramolecular Charge Transfer
References
- Shimomura, O.; Yampolsky, I.V. Bioluminescence: Chemical Principles and Methods, 3rd ed.; World Scientific: Hackensack, NJ, USA, 2019.
- Schmitter, R.; Njus, D.; Sulzman, F.; Gooch, V.; Hastings, J. Dinoflagellate bioluminescence: A comparative study of in vitro components. J. Cell. Physiol. 1976, 87, 123–134.
- Johnson, C.; Inoue, S.; Flint, A.; Hastings, J. Compartmentalization of algal bioluminescence–autofluorescence of bioluminescent particles in the dinoflagellate Gonyaulax as studied with image-intensified video microscopy and flow cytometry. J. Cell Biol. 1985, 100, 1435–1446.
- Knaust, R.; Urbig, T.; Li, L.; Taylor, W.; Hastings, J. The circadian rhythm of bioluminescence in Pyrocystis is not due to differences in the amount of luciferase: A comparative study of three bioluminescent marine dinoflagellates. J. Phycol. 1998, 34, 167–172.
- Akimoto, H.; Wu, C.; Kinumi, T.; Ohmiya, Y. Biological rhythmicity in expressed proteins of the marine dinoflagellate Lingulodinium polyedrum demonstrated by chronological proteomics. Biochem. Biophys. Res. Commun. 2004, 315, 306–312.
- Valiadi, M.; Iglesias-Rodriguez, M. Understanding bioluminescence in dinoflagellates: How far have we come? Microorganisms 2013, 1, 3–25.
- Seo, K.; Fritz, L. Cell ultrastructural changes correlate with circadian rhythms in Pyrocystis lunula (Pyrrophyta). J. Phycol. 2000, 36, 351–358.
- Okamoto, O.; Liu, L.; Robertson, D.; Hastings, J. Members of a dinoflagellate luciferase gene family differ in synonymous substitution rates. Biochemistry 2001, 40, 15862–15868.
- Fajardo, C.; De Donato, M.; Rodulfo, H.; Martinez-Rodriguez, G.; Costas, B.; Mancera, J.M.; Fernandez-Acero, F.J. New perspectives related to the bioluminescent system in dinoflagellates: Pyrocystis lunula, a case study. Int. J. Mol. Sci. 2020, 21, 1784.
- Topalov, G.; Kishi, Y. Chlorophyll catabolism leading to the skeleton of dinoflagellate and krill luciferins: Hypothesis and model studies. Angew. Chem. Int. Ed. 2001, 40, 3892–3894.
- Yamaguchi, A.; Horiguchi, T. Culture of the heterotrophic dinoflagellate Protoperidinium crassipes (Dinophyceae) with noncellular food items. J. Phycol. 2008, 44, 1090–1092.
- Nakamura, H.; Kishi, Y.; Shimomura, O.; Morse, D.; Hastings, J. Structure of dinoflagellate luciferin and its enzymic and nonenzymic air-oxidation products. J. Am. Chem. Soc. 1989, 111, 7607–7611.
- Wang, M.; Liu, Y. Theoretical study of dinoflagellate bioluminescence. Photochem. Photobiol. 2017, 93, 511–518.
- Hamman, J.; Seliger, H. The mechanical triggering of bioluminescence in marine dinoflagellates: Chemical basis. J. Cell. Physiol. 1972, 80, 397–408.
- Dunlap, J. C.; Hastings, W.; Shimomura, O. Cross-reactivity between the light-emitting systems of distantly related organisms: Novel type of light-emitting compound. Proc. Natl. Sci. USA 1980, 77, 1394-1397.
- Liu, L.; Hastings, J. Two different domains of the luciferase gene in the heterotrophic dinoflagellate Noctiluca scintillans occur as two separate genes in photosynthetic species. Proc. Natl. Acad. Sci. USA 2007, 104, 696–701.
- Buskey, E. J.; Strom, S.; Coulter, C. Bioluminescence of heterotrophic dinoflagellates from Texas coastal waters. J. Exp. Mar. Biol. Ecol. 1992, 159, 37-49.
- Wu, C.; Akimoto, H.; Ohmiya, Y. Tracer studies on dinoflagellate luciferin with [15N]-glycine and [15N]-L-glutamic acid in the dinoflagellate Pyrocystis lunula. Tetrahedron Lett. 2003, 44, 1263–1266.
- Dunlap, J.; Hastings, J. Biochemistry of dinoflagellate bioluminescence: Purification and characterization of dinoflagellate luciferin from Pyrocystis lunula. Biochemistry 1981, 20, 983–989.
- Fresneau, C.; Arrio, B. Pyrocystis lunula bioluminescence: Physicochemical characterization of the luciferin precursor. Arch. Biochem. Biophys. 1988, 265, 22–27.
- Fresneau, C.; Hill, M.; Lescure, N.; Arrio, B.; Dupaix, A.; Volfin, P. Dinoflagellate luminescence: Purification of a NAD(P)H-dependent reductase and of its substrate. Arch. Biochem. Biophys. 1986, 251, 495–503.
- Bennoun, P. Evidence for a respiratory chain in the chloroplast. Proc. Natl. Acad. Sci. USA 1982, 79, 4352–4356.
- Peschek, G. The cytochrome oxidase-hydrogenase relationship in cyanobacteria. Naturwissensch 1982, 69, 599–600.
- Omata, T.; Murata, N. Electron-transport reactions in cytoplasmic and thylakoid membranes prepared from the cyanobacteria (blue-green algae) Anacystis nidulans and Synechocystis PCC 6714. Biochim. Biophys. Acta 1985, 810, 354–361.
- Ngo, P.; Mansoorabadi, S. Investigation of the Dinoflagellate Bioluminescence Mechanism: Chemically Initiated Electron Exchange Luminescence or Twisted Intramolecular Charge Transfer? ChemPhotoChem 2017, 1, 383–387.
- Wilson, T.; Hastings, J. Bioluminescence. Ann. Rev. Cell Dev. Biol. 1998, 14, 197–230.
- Koo, J.; Schmidt, S.; Schuster, G. Bioluminescence of the firefly: Key steps in the formation of the electronically excited state for model systems. Proc. Natl. Acad. Sci. USA 1978, 75, 30–33.
- Vacher, M.; Galvaán, I.; Ding, B.W.; Schramm, S.; Berraud-Pache, R.; Naumov, P.; Ferreé, N.; Liu, Y.J.; Navizet, I.; Roca-Sanjuaán, D.; et al. Chemi-and bioluminescence of cyclic peroxides. Chem. Rev. 2018, 118, 6927–6974.
- Grabowski, Z.; Rotkiewicz, K.; Rettig, W. Structural changes accompanying intramolecular electron transfer: Focus on twisted intramolecular charge-transfer states and structures. Chem. Rev. 2003, 103, 3899–4032.
- Janouskovec, J.; Gavelis, G.; Burki, F.; Dinh, D.; Bachvaroff, T.; Gornik, S.; Bright, K.; Imanian, B.; Strom, S.; Delwiche, C.; et al. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc. Natl. Acad. Sci. USA 2017, 114, E171–E180.
- Gornik, S.; Febrimarsa; Cassin, A.M.; MacRae, J.; Ramaprasad, A.; Rchiad, Z.; McConville, M.; Bacic, A.; McFadden, G.; Pain, A.; et al. Endosymbiosis undone by stepwise elimination of the plastid in a parasitic dinoflagellate. Proc. Natl. Acad. Sci. USA 2015, 112, 5767–5772.
- Martin, W.F.; Bryant, D.A.; Beatty, J.T. A physiological perspective on the origin and evolution of photosynthesis. FEMS Microbiol. Rev. 2018, 42, 205–231.
- Fujita, Y.; Tsujimoto, R.; Aoki, R. Evolutionary aspects and regulation of tetrapyrrole biosynthesis in cyanobacteria under aerobic and anaerobic environments. Life 2015, 5, 1172–1203.
- Okamoto, O.; Hastings, J. Genome-wide analysis of redox-regulated genes in a dinoflagellate. Gene 2003, 4, 73–81.
- Buchanan, B. Thioredoxin System and Glutaredoxin Systems; Raven Press: New York, NY, USA, 1985.
- Allocati, N.; Federici, L.; Masulli, M.; Di Ilio, C. Glutathione transferases in bacteria. FEBS J. 2009, 276, 58–75.
- Atkinson, H.; Babbitt, P. Glutathione transferases are structural and functional outliers in the thioredoxin fold. Biochemistry 2009, 48, 11108–11116.
- Hartley, D.; Ruth, J.; Petersen, D. The hepatocellular metabolism of 4-hydroxynonenal by alcohol dehydrogenase, aldehyde dehydrogenase and glutathione S-transferase. Arch. Biochem. Biophys. 1995, 16, 197–205.
- Yamauchi, Y.; Hasegawa, A.; Taninaka, A.; Mizutani, M.; Sugimoto, Y. NADPH-dependent reductases involved in the detoxification of reactive carbonyls in plants. J. Biol. Chem. 2011, 286, 6999–7009.
- Fajardo, C.; Amil-Ruiz, F.; Fuentes-Almagro, C.; De Donato, M.; Martínez-Rodriguez, G.; Escobar-Niño, A.; Carrasco, R.; Mancera, J.M.; Fernandez-Acero, F.J. An “omic” approach to Pyrocystis lunula: New insights related with this bioluminescent dinoflagellate. J. Proteom. 2019, 209, 103502.
- Yasgar, A.; Shultz, J.; Zhou, W.; et al. A high-throughput 1,536-well luminescence assay for glutathione S-transferase activity. Assay Drug Dev. Technol. 2010, 8(2), 200-211.
- Zhou, W.; Shultz, J.W.; Murphy, N.; Hawkins, E.M.; Bernad, L.; Good, T.; et al. Electrophilic aromatic substituted luciferins as bioluminescent probes for glutathione S-transferase assays. Chem. Commun. (Camb) 2006, 4620 – 4622.
- Branchini, B.R.; Hayward, M.M.; Bamford, S.; Brennan, P.M.; Lajiness, E.J. Naphthyl and quinolylluciferin: green and red light emitting firefly luciferin analogues. Photochem. Photobiol. 1989, 49, 689 – 695.
- Beigi, R.; Kobatake, E.; Aizawa, M.; Dubyak, GR. Detection of local ATP release from activated platelets using cell surface-attached firefly luciferase. Am. J. Physiol. Cell Physiol. 1999, 276, 267–278.
- Niwa, K.; Nakajima, Y.; Ohmiya, Y. Applications of luciferin biosynthesis: Bioluminescence assays for L-cysteine and luciferase. Anal. Biochem. 2009, 396(2), 316-318.
- Branchini, BR.; Southworth TL. A highly sensitive biosensor for ATP using a chimeric firefly luciferase. Methods Enzymol. 2017, 589, 351-364.
- Branchini, B.R.; Southworth, T.L.; Fontaine, D.M.; Kohrt, D.; Welcome, F.S.; Florentine, C.M.; Henricks, E.R.; DeBartolo, D.B.; Michelini, E.; Cevenin,i L.; Roda, A.; Grossel, M.J. Red-emitting chimeric firefly luciferase for in vivo imaging in low ATP cellular environments. Anal. Biochem. 2017, 534, 36-39.
- Minekawa, T.; Ohkuma, H.; Abe, K.; Maekawa, H.; Arakawa, H. Practical application of bioluminescence enzyme immunoassay using enhancer for firefly luciferin-luciferase bioluminescence. Luminescence 2011, 26(3), 167-71.
- Marques, S.M.; Esteves da Silva, J.C.G. Firefly bioluminescence: A mechanistic approach of luciferase catalyzed reactions. IUBMB Life 2009, 61: 6-17.
- Branchini, B.R.; Southworth, T.L.; Fontaine, D.M. et al. A firefly luciferase dual color bioluminescence reporter assay using two substrates to simultaneously monitor two gene expression events. Sci. Rep. 2018, 8, 5990.
- Yuma Ikeda, Y.; Saitoh, T.; Niwa, K.; Nakajima, T.; Kitada, N.; Shojiro, A.; Maki; Sato, M.; Citterio, D.; Nishiyamaf, S.; Suzuki, K. An allylated firefly luciferin analogue with luciferase specific response in living cells. Chem. Comm. 2018, 14.
- Podsiadły, R.; Grzelakowska, A.; Modrzejewska, J.; Siarkiewicz, P.; Słowiński, D.; Szala, M.; Świerczyńska, M. Recent progress in the synthesis of firefly luciferin derivatives. Dyes and Pigments 2019, 170, 107627.
- Yuma Ikeda; Takahiro Nomoto; Yuki Tokura; Nobuhiro Nishiyama; Daniel Citterio; Ring-Fused Firefly Luciferins: Expanded Palette of Near-Infrared Emitting Bioluminescent Substrates. Analytical Chemistry 2020, 92, 4235-4243, 10.1021/acs.analchem.9b04562.
- Yuma Ikeda; Takahiro Nomoto; Yuki Tokura; Nobuhiro Nishiyama; Daniel Citterio; Ring-Fused Firefly Luciferins: Expanded Palette of Near-Infrared Emitting Bioluminescent Substrates. Analytical Chemistry 2020, 92, 4235-4243, 10.1021/acs.analchem.9b04562.