Amino acids have been extensively studied in nutrition, mainly as key elements for maintaining optimal protein synthesis in the body as well as precursors of various nitrogen-containing compounds. However, it is now known that amino acid catabolism is an important element for the metabolic control of different biological processes, although it is still a developing field to have a deeper understanding of its biological implications. The mechanisms involved in the regulation of amino acid catabolism now include the contribution of the gut microbiota to amino acid oxidation and metabolite generation in the intestine, the molecular mechanisms of transcriptional control, and the participation of specific miRNAs involved in the regulation of amino acid degrading enzymes. In addition, molecules derived from amino acid catabolism play a role in metabolism as they are used in the epigenetic regulation of many genes.
- amino acid catabolism
- gut microbiota
- gene transcription
- epigenetics
- immune response
- obesity
- diabetes
- thermogenesis
1. Introduction
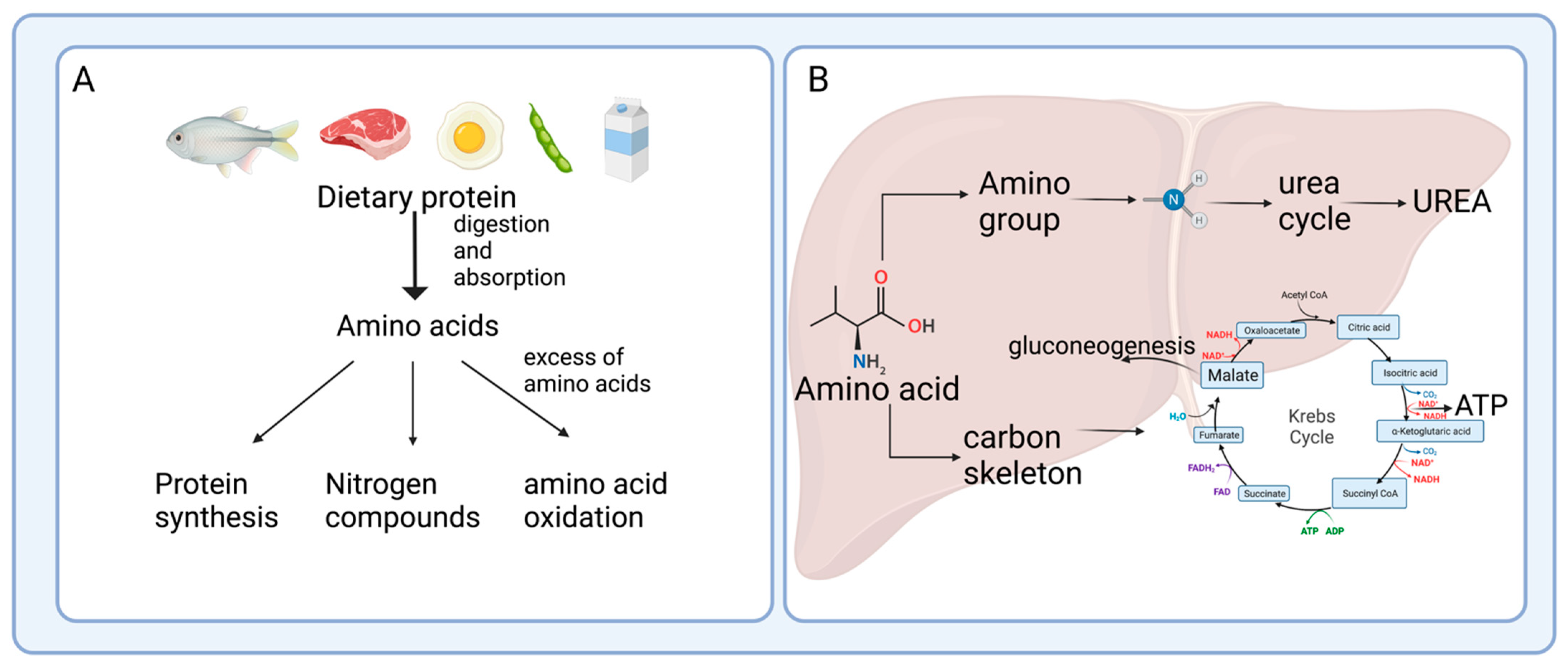
2. Enterohepatic Axis of Amino Acid Catabolism
After protein digestion, amino acids are absorbed and used not only by intestinal cells but also by the resident bacteria; once they have passed through the gut, a fraction of the amino acids are transferred to the bloodstream to reach the liver [4]. Several studies have shown active amino acid utilization and turnover in the enterohepatic axis, which is key for several physiological processes, including the immune response [5], bacteria metabolism, and generation of end-products, where the intestine and the gut microbiota play an important role [6].2.1. Protein Hydrolysis and Amino Acid Absorption in the Gut
After passing through the esophagus and gastric lumen, dietary protein is hydrolyzed in the small intestine by the proteases and peptidases, including trypsin, chymotrypsin, or carboxypeptidases released by the pancreas [7]. In the intestinal mucosa, an intense renewal of protein occurs at approximately 50% per day in humans, indicating that amino acids are key effectors of gut protein turnover [8]. Many studies have focused on two amino acids, glutamine and arginine, in protein synthesis; however, amino acid catabolism in the intestine has not been extensively documented [8]. The brush border cells of the intestinal mucosa release some peptidases that are used for protein hydrolysis; these enzymes are more active at neutral to alkaline pH, which is reached in the small intestine, to catalyze the hydrolysis of dietary protein to produce amino acids and peptides that are actively absorbed by the enterocytes [9]. Although the process is quite efficient, some nitrogenated products are not taken up by the small intestine, pass through the ileocecal junction, and can be found in the large intestine [10]. Figure 2A outlines this process of protein digestion and absorption of peptides and amino acids, showing its passage through the gastrointestinal system.2.2. Amino Acid Catabolism in the Intestine
Amino acids and some peptides found in the intestinal lumen after digestion can be delivered to the portal vein due to the presence of specific transporters known as solute carriers (SCL) in the brush border or apical membrane (SLC1A1, SLC6A19, SLC7A1, SLC38A5, SLC36A1, SLC15A1 transporters), and in the basolateral membrane of the enterocytes (members of the SLC7A family and SCL16A10, SLC38A2 transporters) [11] (Figure 2B). However, a large number of dietary amino acids, particularly BCAA, are partially catabolized in the gut since branched-chain aminotransferase (BCAT), the first enzyme in the degradation of the BCAA leucine, valine, and isoleucine, has been reported to be present in the gut [12]. Almost all dietary glutamate, aspartate, and approximately 30–70% of BCAA, glutamine, proline, lysine, threonine, methionine, and phenylalanine are metabolized in the small intestine of mammals, including humans [13]. These data suggest that several amino acid degrading enzymes (AADEs) are expressed in enterocytes. It has been shown that cells of the small intestinal mucosa express lysine α-ketoglutarate reductase in pigs [14]; methionine transamination [15] and glutamine transaminase K in rats [16]; threonine dehydrogenase in pigs [17]; BCAA transaminase and branched-chain α-keto acid (BCKA) dehydrogenase in humans and pigs [2][18]; and proline oxidase in pigs and rats [19]. Also, several enzymes for arginine and glutamine degradation are present, including arginase II, phosphate-dependent glutaminase (PDG), carbamoylphosphate synthase II (glutamine) (CPS-II), glutamate-oxaloacetate transaminase (GOT), glutamate-pyruvate transaminase (GPT), pyrroline-5-carboxylate (P5C) synthase, ornithine aminotransferase (OAT), P5C reductase, ornithine carbamoyltransferase (OCT), carbamoylphosphate synthase I (ammonia) (CPS-I), argininosuccinate synthase (ASS), argininosuccinate lyase (ASL), and ornithine decarboxylase (ODC) [20][21][22]. In the colonic lumen, colonocytes can use glutamine as a fuel substrate and catabolize arginine into ornithine and nitric oxide [23]. The conversion of arginine into ornithine significantly increases its capacity in response to an elevated protein intake, which attenuates the rising ammonia concentration in the blood if the urea cycle requires more ornithine [24].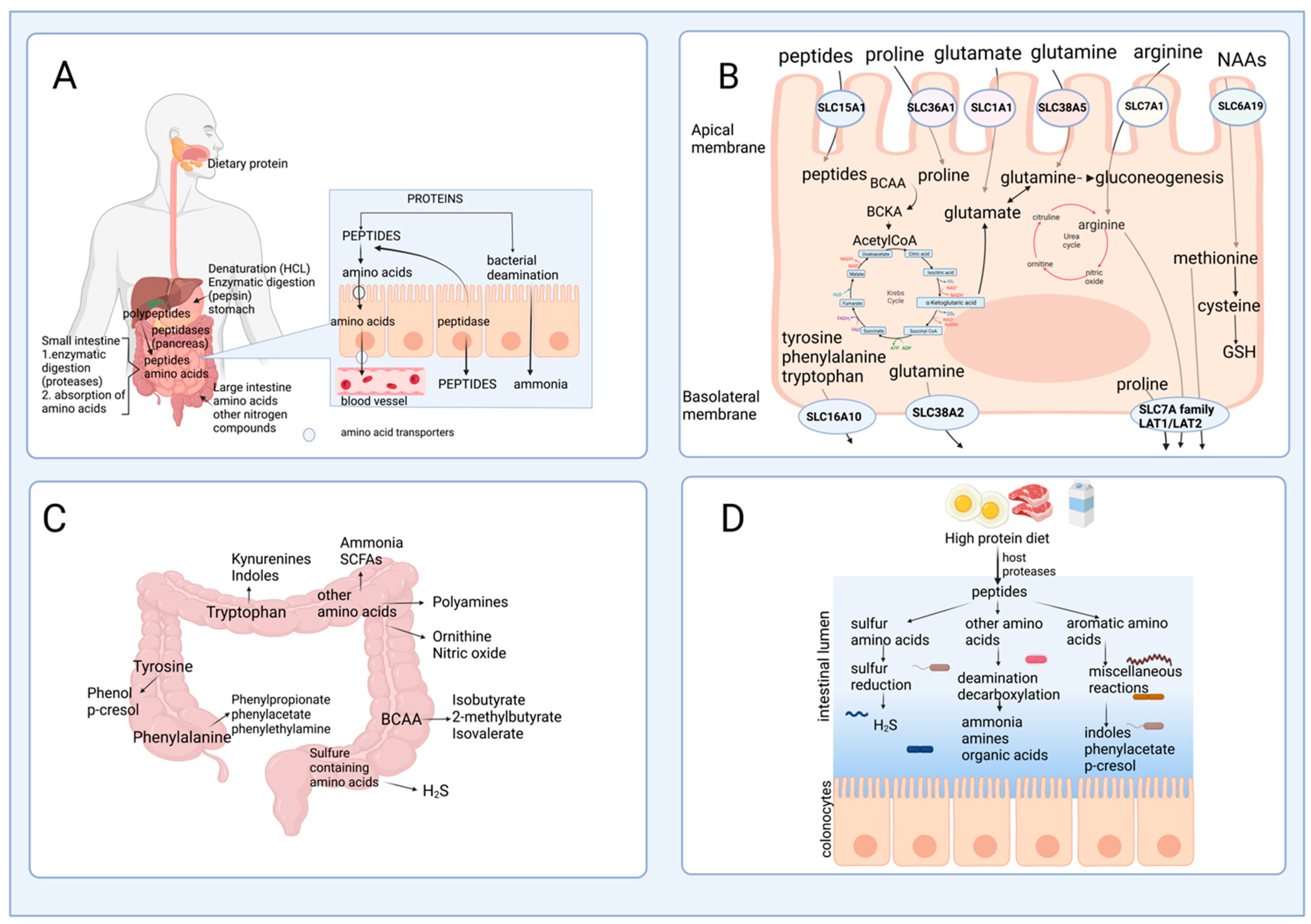
2.3. The Role of Gut Microbiota in Amino Acid Catabolism and Generation of End-Products
Studies in germ-free animals reveal that gut bacteria alter the distribution of free amino acids in the gastrointestinal tract, indicating the role of the bacteria in the synthesis or degradation of amino acids. Interestingly, the amount of dietary protein and the dietary protein source modify the gut microbiota [26]. Consequently, this may suggest that the gut microbiota affects the bioavailability of amino acids to the host, depending on the taxonomy of intestinal bacteria. In addition, although short-chain fatty acids (SCFA) are the primary end product of carbohydrate fermentation, many amino acids formed by reductive deamination by bacteria can be precursors of SCFA. Certain bacteria in the small intestine, including Klebsiella spp., Escherichia coli, Streptococcus spp., Succinivibrio dextrinosolvens, Mitsuokella spp., and Anaerovibrio lipolytica, can also utilize protein digestive products and play an essential role in nitrogen recycling [27][28]. Moreover, bacteria species such as Prevotella ruminicola, Butyrivibrio fibrisolvens, Mitsuokella multiacidas, and Streptococcus bovis can secrete active dipeptidyl peptidase and dipeptidase that contribute to protein digestion and absorption of amino acids in the gut [29]. Ammonia is released daily by bacteria through the deamination of amino acids and, to a lesser extent, through urea hydrolysis catalyzed by bacterial urease activity. Up to 3.5–4.0 g of ammonia are released daily in the gut. Ammonia can be used by the bacteria for their metabolism and protein synthesis, absorbed by the colonocytes, transformed into urea in the liver, and excreted in urine [30]. It has been suggested that bacteria in the small intestine may participate in the catabolism of some essential amino acids and modulate the bioavailability of amino acids and their end-products in the systemic circulation of animals and humans. A previous study using conventional and germ-free rats with 15N incorporation into body lysine demonstrated that the N-lysine measured in the host came mainly from bacteria, indicating that amino acids can be exchanged between the host and the microbiota [31]. Although the digestion of dietary protein followed by amino acid absorption is a very efficient process in the small intestine, some nitrogenous material may escape digestion and be transferred into the large intestine. Undigested peptides and amino acids are usually not absorbed by the colonocytes but are fermented by gut microbes into intermediate metabolites and other end-products (Figure 2C), and this will depend on the amount of dietary protein consumed (Figure 2D). Some bacteria genera with proteolytic activity in the large intestine are Bacteroides, Propionibacterium, Streptococcus, Fusobacterium, Clostridium, and Lactobacillus [10][32]. Bacterial degradation of aromatic amino acids in the colonic lumen results in the production of phenolic and indolic compounds, ammonia, polyamines, and hydrogen sulfide [33][34]. Moreover, the catabolism of amino acids in these cells produces multiple metabolites, such as p-cresol from tyrosine, phenylpropionate, phenylacetate from phenylalanine, and indole skatole from tryptophan [30]. Phenolic compounds are primarily absorbed from the colon, detoxified in the colon mucosa and the liver by glucuronide and sulfate conjugation, and excreted in the urine. Decarboxylation of amino acids results in the appearance of amines in the gut. Monoamine and diamine oxidases present in the gut mucosa detoxify the amines produced by the gut microbiota [30].3. Mechanisms of Regulation of Amino Acid Oxidation
Amino acid catabolism involves several metabolic pathways specific to a particular group of amino acids, involving multiple enzymes, some of which are essential for regulating the flux of metabolites in these pathways and are known as amino acid degrading enzymes (AADEs). AADEs are mainly located in the liver but are now known to be found in other extrahepatic tissues; they are specific for the utilization of each amino acid, and the transcription of the genes encoding them is highly regulated [35]. The study of the molecular mechanisms of regulation of AADEs had a significant breakthrough in the late 1980s and during the 1990s, when the transcriptional regulation of many AADEs by diet and various hormones was elucidated. These investigations provided sufficient information to understand certain key aspects of transcriptional control of AADEs that we now know are highly conserved among the genes of AADEs; however, recent studies have enriched previous knowledge by involving the participation of new players on the scene.4. Amino Acid Catabolism in Health and Disease
Amino acid catabolism provides metabolites that serve as substrates or are responsible for activating signaling pathways involved in the physiology of different organs. For example, the regulation of amino acid catabolism is key for the immune system since its functions depend on an adequate supply of amino acids, and individual amino acids and their metabolites can affect immune responses [5]. Similarly, amino acid catabolism dysregulation can contribute to the development, or progression of pathological processes involved in the presence of insulin resistance [36]. Here wthe researchers provide evidence for the importance of amino acid turnover and catabolism in health and disease, opening the landscape to new mechanisms regulated by amino acids and their metabolites beyond their known functions for protein synthesis and energy fuel.4.1. Amino Acid Catabolism and Immunity
All cells, including those involved in immune responses, depend on nutrient availability to maintain their functionality [37]. When there is an inflammatory or antigenic cue, immune cells need more amino acids to remain viable and respond accordingly, so they must adapt rapidly to any shortage of amino acids [38]. This adaptation suggests that any modification in amino acid (and other nutrient) metabolism will also affect the immune response in different ways, depending on the specific nutrient and energy requirements of the cells and, thus, their function [39][40]. In other words, each immune cell’s microenvironment will be directly related to their response to nutrient availability. Both the innate and adaptive immune systems require an adequate supply of amino acids to synthesize molecules such as histamine, glutathione, and nitric oxide, among others, but especially for immunoglobulins and cytokine activation, as well as for antibody production through mTOR signaling by BCAA [41]. Therefore, individual amino acids and their metabolites can affect immune responses. Besides BCAA, the AADE that have been mainly studied for their response to inflammation are those involved in tryptophan and arginine catabolism, such as tryptophan 2,3-dioxygenase (TDO), IDO1, the arginase isoforms (ARG1, ARG2), and the inducible nitric oxide synthase (iNOS), respectively [42].4.2. Amino Acid Catabolism in Obesity and Diabetes
An adequate amino acid catabolism is crucial to maintaining plasma amino acid concentration. Disturbances in plasma amino acid concentration have been associated with alterations in individual health [36]. In a cross-sectional study with young adults, it has been observed that subjects with obesity have higher levels of alanine, aspartate, cysteine, ornithine, phenylalanine, proline, and tyrosine and lower levels of glycine, ornithine, and serine compared to normal weight subjects [43]. Furthermore, subjects with insulin resistance (IR) (defined as HOMA > 2.5) have higher levels of arginine, alanine, aspartate, isoleucine, leucine, phenylalanine, proline, tyrosine, taurine, and valine than subjects without IR [43]. In addition, increased levels of BCAA and aromatic amino acids are associated with a five-fold increased risk of developing type 2 diabetes [44][45]. BCAAs play important general roles in the body, including regulation of protein synthesis through mTOR and as a source of energy during exercise, which have been extensively reviewed elsewhere [46]. However, there is significant controversy as to whether altered plasma concentrations of amino acids, especially BCAA, are a cause or consequence of obesity or insulin resistance. Unlike the rest of the amino acids, as mentioned above, the first enzymatic reaction of BCAA catabolism is extrahepatic, with muscle, kidney, and adipose tissue as the principal organs for BCAA transamination. Notably, BCAA catabolism is mainly impaired in adipose tissue during obesity. Branched-chain aminotransferase 2 (BCAT2), an isoform of BCAT found in mitochondria, and BCKDH activity are reduced in the adipose tissue of mice and rats with genetic or diet-induced obesity [47][48][49][50]. In subjects with obesity, BCAT2 and BCKDH expression is reduced mainly in visceral adipose tissue [48][51]. Nevertheless, further research is needed to clarify the mechanisms responsible for the decreased expression and activity of BCAT2 and BCKDH in adipose tissue during obesity. The alteration of BCAA catabolism has physiological consequences in two aspects. First, leucine is likely the most potent mTORC1 activator among all amino acids. In physiological conditions, mTORC1 activation inhibits autophagy, activates adipogenesis and lipogenesis in white adipose tissue, and inhibits insulin signaling through the inactivation of the insulin receptor substrate 1 (IRS1). Thus, high leucine concentrations could induce an overactivation of mTORC1, leading to insulin resistance and, thus, an increase in glucose levels [52][53]. The use of rapamycin, a known mTORC1 inhibitor, prevents glucose intolerance induced by a diet high in fat and BCAA [54]. Furthermore, leucine catabolism contributes 30% to the acetyl-CoA lipogenic pool [55]. and acetyl-CoA can be converted into malonyl-CoA by acetyl-CoA carboxylase (ACC). Malonyl-CoA is the preferred substrate of fatty acid synthase (FAS). Thus, leucine is extensively incorporated into the lipid fraction of functional adipocytes. Incorporation that is significantly reduced in adipocytes from high-fat fed rats [49]. In addition to fatty acids, the carbon skeleton of leucine could be a substrate for synthesizing phospholipids and cholesterol. An increase in cholesterol-synthetizing enzymes has been demonstrated, which use 3-hydroxy-3-methylglutaryl-CoA, an intermediate of leucine catabolism, as a substrate during adipogenesis [56]. Taken together, this evidence suggests that decreased catabolism of BCAA could reduce the production and storage of FA and cholesterol in adipose tissue, affecting its functionality. Second, a reduction in the BCAA catabolic enzymes causes an inefficient production of anaplerotic substrates from BCAA, causing suboptimal activity of the Krebs cycle [57][58]. These results have important implications for the understanding of metabolic inflexibility. Until now, metabolic flexibility has only been assessed in terms of how glucose affects lipid metabolism and vice versa. However, new evidence suggests that amino acid catabolism needs to be evaluated in terms of metabolic flexibility because amino acid availability and catabolic amino acid metabolites can also affect lipid and carbohydrate metabolism, as in the model proposed by Muoio, where chronic overfeeding causes a mitochondrial blockade affecting glucose, fatty acid, and BCAA oxidation [59]. Although progress has been made in understanding the participation of BCAA catabolism in the maintenance of adipocyte function, further research is needed to understand the role of catabolism of other amino acids such as aspartate, proline, tyrosine, and tryptophan, among others, that can modify mitochondrial activity and therefore metabolic flexibility in the adipocyte. In addition to BCAAs, tryptophan (Trp) is another essential amino acid for humans, [60]. Consumed tryptophan is mainly used for protein synthesis; however, this amino acid can be used in about 5% of cases as a precursor for the synthesis of serotonin, N-acetyl serotonin, and melatonin [61]. Interestingly, a part of free tryptophan is also catabolized through the tryptophan-kynurenine pathway [62]. Recent studies have found that certain metabolites of tryptophan catabolism participate in the development of T2D [63]. A significant association between low plasma Trp concentrations has been reported in obese subjects with metabolic syndrome, in whom insulin resistance is common [64]. Evidence suggests that metabolites of the kynurenine pathway increase with insulin resistance before the clinical manifestation of hyperglycemia [65]. Gene expression of tryptophan catabolism limiting enzymes to kynurenine, such as indolamine 2,3-dioxygenase 1 (IDO1), indolamine 2,3-dioxygenase 2 (IDO2), and tryptophan 2,3-dioxygenase (TDO2), is shown to increase in patients with T2D [66]. A possible mechanism by which metabolites of the Trp-Kynurenine pathway contribute to the development of insulin resistance includes the possible formation of chelate complexes between xanthurenic acid and insulin, which are indistinguishable from free insulin but have ∼50% less activity than insulin [67]. However, studies are still needed to determine the importance of the regulation of gene expression by the step-limiting enzyme of the Trp- Kynurenine pathway in the pathogenesis of diabetes or its complications. On the other hand, Trp and phenylalanine interact with the GPR142 receptor, increasing insulin secretion and the incretins GIP and GLP-1, improving circulating glucose levels [68], suggesting that the decrease in Trp during obesity will decrease insulin secretion and contribute to the progression to T2D of subjects with obesity.4.3. Amino Acid Catabolism and Thermogenesis
In addition to white adipose tissue, it has been demonstrated that BAT may play an essential role in the control of body thermogenesis [69]. This effect is in part mediated by the presence of the uncoupling protein 1 (UCP1), which can use the proton gradient generated in the mitochondria by the respiratory chain in order to produce heat [70]. The energy sources used by BAT mitochondria are fatty acids and glucose [71]. Interestingly, recent evidence has demonstrated that BAT mitochondria can use BCAA amino acids as an energy source to increase thermogenesis. BAT is now known to express enzymes of the amino acid catabolic pathways for BCAA, including BCAT2 and BCKDH. Under conditions of cold exposure, where thermogenesis is upregulated, there is a significantly increased uptake of BCAA by BAT, particularly valine and leucine. The importance of BCAA oxidation for thermogenesis has been demonstrated since the absence of BCKDH expression in BAT impairs energy homeostasis, especially during cold exposure. BCAA oxidation occurs in the mitochondria, and it is known that BCAAs are transported into the mitochondria by the SLC25A44 transporter. Surprisingly, the deletion of SLC25A44 impairs BAT thermogenesis [72]. As previously mentioned, white adipose tissue plays an important role in the utilization of BCAAs, which is associated with improving insulin sensitivity. However, now BAT is also considered an essential catabolic organ of these amino acids, decreasing circulating levels of BCAA that are associated with increased insulin sensitivity.References
- Chou, C.J.; Affolter, M.; Kussmann, M. A nutrigenomics view of protein intake: Macronutrient, bioactive peptides, and protein turnover. Prog. Mol. Biol. Transl. Sci. 2012, 108, 51–74.
- Harper, A.E.; Miller, R.H.; Block, K.P. Branched-chain amino acid metabolism. Annu. Rev. Nutr. 1984, 4, 409–454.
- Bender, D.A. The metabolism of “surplus” amino acids. Br. J. Nutr. 2012, 108 (Suppl. S2), S113–S121.
- Bartlett, A.; Kleiner, M. Dietary protein and the intestinal microbiota: An understudied relationship. iScience 2022, 25, 105313.
- Tome, D. Amino acid metabolism and signalling pathways: Potential targets in the control of infection and immunity. Nutr. Diabetes 2021, 11, 20.
- Broer, S. Amino acid transport across mammalian intestinal and renal epithelia. Physiol. Rev. 2008, 88, 249–286.
- Loveday, S.M. Protein digestion and absorption: The influence of food processing. Nutr. Res. Rev. 2022, 16, 1–16.
- Bertrand, J.; Goichon, A.; Dechelotte, P.; Coeffier, M. Regulation of intestinal protein metabolism by amino acids. Amino Acids 2013, 45, 443–450.
- Sleisenger, M.H.; Kim, Y.S. Protein digestion and absorption. N. Engl. J. Med. 1979, 300, 659–663.
- Davila, A.M.; Blachier, F.; Gotteland, M.; Andriamihaja, M.; Benetti, P.H.; Sanz, Y.; Tome, D. Intestinal luminal nitrogen metabolism: Role of the gut microbiota and consequences for the host. Pharmacol. Res. 2013, 68, 95–107.
- Chen, C.; Yin, Y.; Tu, Q.; Yang, H. Glucose and amino acid in enterocyte: Absorption, metabolism and maturation. Front. Biosci. 2018, 23, 1721–1739.
- Torres, N.; Lopez, G.; De Santiago, S.; Hutson, S.M.; Tovar, A.R. Dietary protein level regulates expression of the mitochondrial branched-chain aminotransferase in rats. J. Nutr. 1998, 128, 1368–1375.
- Ma, N.; Ma, X. Dietary Amino Acids and the Gut-Microbiome-Immune Axis: Physiological Metabolism and Therapeutic Prospects. Compr. Rev. Food Sci. Food Saf. 2019, 18, 221–242.
- Pink, D.B.; Gatrell, S.K.; Elango, R.; Turchinsky, J.; Kiess, A.S.; Blemings, K.P.; Dixon, W.T.; Ball, R.O. Lysine alpha-ketoglutarate reductase, but not saccharopine dehydrogenase, is subject to substrate inhibition in pig liver. Nutr. Res. 2011, 31, 544–554.
- Mitchell, A.D.; Benevenga, N.J. The role of transamination in methionine oxidation in the rat. J. Nutr. 1978, 108, 67–78.
- Cooper, A.J.; Meister, A. Comparative studies of glutamine transaminases from rat tissues. Comp. Biochem. Physiol. B Biochem. 1981, 69, 137–145.
- Schaart, M.W.; Schierbeek, H.; van der Schoor, S.R.; Stoll, B.; Burrin, D.G.; Reeds, P.J.; van Goudoever, J.B. Threonine utilization is high in the intestine of piglets. J. Nutr. 2005, 135, 765–770.
- Hoerr, R.A.; Matthews, D.E.; Bier, D.M.; Young, V.R. Effects of protein restriction and acute refeeding on leucine and lysine kinetics in young men. Am. J. Physiol. 1993, 264, E567–E575.
- Wu, G. Synthesis of citrulline and arginine from proline in enterocytes of postnatal pigs. Am. J. Physiol. 1997, 272, G1382–G1390.
- Bush, J.A.; Wu, G.; Suryawan, A.; Nguyen, H.V.; Davis, T.A. Somatotropin-induced amino acid conservation in pigs involves differential regulation of liver and gut urea cycle enzyme activity. J. Nutr. 2002, 132, 59–67.
- Morris, S.M., Jr. Regulation of enzymes of the urea cycle and arginine metabolism. Annu. Rev. Nutr. 2002, 22, 87–105.
- Wu, G.; Morris, S.M., Jr. Arginine metabolism: Nitric oxide and beyond. Biochem. J. 1998, 336 Pt 1, 1–17.
- Kan, C.C.; Chung, T.Y.; Juo, Y.A.; Hsieh, M.H. Glutamine rapidly induces the expression of key transcription factor genes involved in nitrogen and stress responses in rice roots. BMC Genom. 2015, 16, 731.
- Mouille, B.; Robert, V.; Blachier, F. Adaptative increase of ornithine production and decrease of ammonia metabolism in rat colonocytes after hyperproteic diet ingestion. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 287, G344–G351.
- Piper, D.W.; Fenton, B.H. pH stability and activity curves of pepsin with special reference to their clinical importance. Gut 1965, 6, 506–508.
- Sanchez-Tapia, M.; Moreno-Vicencio, D.; Ordaz-Nava, G.; Guevara-Cruz, M.; Granados-Portillo, O.; Vargas-Castillo, A.; Torres, N.; Tovar, A.R. Antibiotic Treatment Reduces the Health Benefits of Soy Protein. Mol. Nutr. Food Res. 2020, 64, e2000532.
- Dai, Z.L.; Li, X.L.; Xi, P.B.; Zhang, J.; Wu, G.; Zhu, W.Y. Metabolism of select amino acids in bacteria from the pig small intestine. Amino Acids 2012, 42, 1597–1608.
- Dai, Z.L.; Zhang, J.; Wu, G.; Zhu, W.Y. Utilization of amino acids by bacteria from the pig small intestine. Amino Acids 2010, 39, 1201–1215.
- Fan, P.; Liu, P.; Song, P.; Chen, X.; Ma, X. Moderate dietary protein restriction alters the composition of gut microbiota and improves ileal barrier function in adult pig model. Sci. Rep. 2017, 7, 43412.
- Windey, K.; De Preter, V.; Verbeke, K. Relevance of protein fermentation to gut health. Mol. Nutr. Food Res. 2012, 56, 184–196.
- Torrallardona, D.; Harris, C.I.; Coates, M.E.; Fuller, M.F. Microbial amino acid synthesis and utilization in rats: Incorporation of 15N from 15NH4Cl into lysine in the tissues of germ-free and conventional rats. Br. J. Nutr. 1996, 76, 689–700.
- Macfarlane, G.T.; Allison, C.; Gibson, S.A.; Cummings, J.H. Contribution of the microflora to proteolysis in the human large intestine. J. Appl. Bacteriol. 1988, 64, 37–46.
- Blachier, F.; Mariotti, F.; Huneau, J.F.; Tome, D. Effects of amino acid-derived luminal metabolites on the colonic epithelium and physiopathological consequences. Amino Acids 2007, 33, 547–562.
- Macfarlane, G.T.; Gibson, S.A.; Beatty, E.; Cummings, J.H. Estimation of short-chain fatty acid production from protein by human intestinal bacteria based on branched-chain fatty acid measurements. FEMS Microbiol. Lett. 1992, 101, 81–88.
- Desvergne, B.; Michalik, L.; Wahli, W. Transcriptional regulation of metabolism. Physiol. Rev. 2006, 86, 465–514.
- Vanweert, F.; Schrauwen, P.; Phielix, E. Role of branched-chain amino acid metabolism in the pathogenesis of obesity and type 2 diabetes-related metabolic disturbances BCAA metabolism in type 2 diabetes. Nutr. Diabetes 2022, 12, 35.
- Newsholme, P. Cellular and metabolic mechanisms of nutrient actions in immune function. Eur. J. Clin. Nutr. 2021, 75, 1328–1331.
- Kelly, B.; Pearce, E.L. Amino Assets: How Amino Acids Support Immunity. Cell Metab. 2020, 32, 154–175.
- Kedia-Mehta, N.; Finlay, D.K. Competition for nutrients and its role in controlling immune responses. Nat. Commun. 2019, 10, 2123.
- Loftus, R.M.; Finlay, D.K. Immunometabolism: Cellular Metabolism Turns Immune Regulator. J. Biol. Chem. 2016, 291, 1–10.
- Calder, P.C. Branched-chain amino acids and immunity. J. Nutr. 2006, 136, 288S–293S.
- McGaha, T.L.; Huang, L.; Lemos, H.; Metz, R.; Mautino, M.; Prendergast, G.C.; Mellor, A.L. Amino acid catabolism: A pivotal regulator of innate and adaptive immunity. Immunol. Rev. 2012, 249, 135–157.
- Guevara-Cruz, M.; Vargas-Morales, J.M.; Mendez-Garcia, A.L.; Lopez-Barradas, A.M.; Granados-Portillo, O.; Ordaz-Nava, G.; Rocha-Viggiano, A.K.; Gutierrez-Leyte, C.A.; Medina-Cerda, E.; Rosado, J.L.; et al. Amino acid profiles of young adults differ by sex, body mass index and insulin resistance. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 393–401.
- Medina-Vera, I.; Sanchez-Tapia, M.; Noriega-Lopez, L.; Granados-Portillo, O.; Guevara-Cruz, M.; Flores-Lopez, A.; Avila-Nava, A.; Fernandez, M.L.; Tovar, A.R.; Torres, N. A dietary intervention with functional foods reduces metabolic endotoxaemia and attenuates biochemical abnormalities by modifying faecal microbiota in people with type 2 diabetes. Diabetes Metab. 2019, 45, 122–131.
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448–453.
- Neinast, M.; Murashige, D.; Arany, Z. Branched Chain Amino Acids. Annu. Rev. Physiol. 2019, 81, 139–164.
- Estrada-Alcalde, I.; Tenorio-Guzman, M.R.; Tovar, A.R.; Salinas-Rubio, D.; Torre-Villalvazo, I.; Torres, N.; Noriega, L.G. Metabolic Fate of Branched-Chain Amino Acids During Adipogenesis, in Adipocytes From Obese Mice and C2C12 Myotubes. J. Cell Biochem. 2017, 118, 808–818.
- Lackey, D.E.; Lynch, C.J.; Olson, K.C.; Mostaedi, R.; Ali, M.; Smith, W.H.; Karpe, F.; Humphreys, S.; Bedinger, D.H.; Dunn, T.N.; et al. Regulation of adipose branched-chain amino acid catabolism enzyme expression and cross-adipose amino acid flux in human obesity. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E1175–E1187.
- Salinas-Rubio, D.; Tovar, A.R.; Torre-Villalvazo, I.; Granados-Portillo, O.; Torres, N.; Pedraza-Chaverri, J.; Noriega, L.G. Interaction between leucine and palmitate catabolism in 3T3-L1 adipocytes and primary adipocytes from control and obese rats. J. Nutr. Biochem. 2018, 59, 29–36.
- She, P.; Van Horn, C.; Reid, T.; Hutson, S.M.; Cooney, R.N.; Lynch, C.J. Obesity-related elevations in plasma leucine are associated with alterations in enzymes involved in branched-chain amino acid metabolism. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E1552–E1563.
- Serralde-Zuniga, A.E.; Guevara-Cruz, M.; Tovar, A.R.; Herrera-Hernandez, M.F.; Noriega, L.G.; Granados, O.; Torres, N. Omental adipose tissue gene expression, gene variants, branched-chain amino acids, and their relationship with metabolic syndrome and insulin resistance in humans. Genes Nutr. 2014, 9, 431.
- Zoncu, R.; Bar-Peled, L.; Efeyan, A.; Wang, S.; Sancak, Y.; Sabatini, D.M. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011, 334, 678–683.
- Zoncu, R.; Efeyan, A.; Sabatini, D.M. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011, 12, 21–35.
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009, 9, 311–326.
- Green, C.R.; Wallace, M.; Divakaruni, A.S.; Phillips, S.A.; Murphy, A.N.; Ciaraldi, T.P.; Metallo, C.M. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nat. Chem. Biol. 2016, 12, 15–21.
- Halama, A.; Horsch, M.; Kastenmuller, G.; Moller, G.; Kumar, P.; Prehn, C.; Laumen, H.; Hauner, H.; Hrabe de Angelis, M.; Beckers, J.; et al. Metabolic switch during adipogenesis: From branched chain amino acid catabolism to lipid synthesis. Arch. Biochem. Biophys. 2016, 589, 93–107.
- Hong, S.; Zhou, W.; Fang, B.; Lu, W.; Loro, E.; Damle, M.; Ding, G.; Jager, J.; Zhang, S.; Zhang, Y.; et al. Dissociation of muscle insulin sensitivity from exercise endurance in mice by HDAC3 depletion. Nat. Med. 2017, 23, 223–234.
- Lerin, C.; Goldfine, A.B.; Boes, T.; Liu, M.; Kasif, S.; Dreyfuss, J.M.; De Sousa-Coelho, A.L.; Daher, G.; Manoli, I.; Sysol, J.R.; et al. Defects in muscle branched-chain amino acid oxidation contribute to impaired lipid metabolism. Mol. Metab. 2016, 5, 926–936.
- Muoio, D.M. Metabolic inflexibility: When mitochondrial indecision leads to metabolic gridlock. Cell 2014, 159, 1253–1262.
- Chandel, N.S. Amino Acid Metabolism. Cold Spring Harb. Perspect. Biol. 2021, 13, a040584.
- Oxenkrug, G.F. Genetic and hormonal regulation of tryptophan kynurenine metabolism: Implications for vascular cognitive impairment, major depressive disorder, and aging. Ann. N. Y. Acad. Sci. 2007, 1122, 35–49.
- Schwarcz, R. The kynurenine pathway of tryptophan degradation as a drug target. Curr. Opin. Pharmacol. 2004, 4, 12–17.
- Chen, T.; Zheng, X.; Ma, X.; Bao, Y.; Ni, Y.; Hu, C.; Rajani, C.; Huang, F.; Zhao, A.; Jia, W.; et al. Tryptophan Predicts the Risk for Future Type 2 Diabetes. PLoS ONE 2016, 11, e0162192.
- Breum, L.; Rasmussen, M.H.; Hilsted, J.; Fernstrom, J.D. Twenty-four-hour plasma tryptophan concentrations and ratios are below normal in obese subjects and are not normalized by substantial weight reduction. Am. J. Clin. Nutr. 2003, 77, 1112–1118.
- Muzik, O.; Burghardt, P.; Yi, Z.; Kumar, A.; Seyoum, B. Successful metformin treatment of insulin resistance is associated with down-regulation of the kynurenine pathway. Biochem. Biophys. Res. Commun. 2017, 488, 29–32.
- Oxenkrug, G.; van der Hart, M.; Summergrad, P. Elevated anthranilic acid plasma concentrations in type 1 but not type 2 diabetes mellitus. Integr. Mol. Med. 2015, 2, 365–368.
- Kotake, Y.; Ueda, T.; Mori, T.; Igaki, S.; Hattori, M. Abnormal tryptophan metabolism and experimental diabetes by xanthurenic acid (XA). Acta Vitaminol. Enzymol. 1975, 29, 236–239.
- Lin, H.V.; Efanov, A.M.; Fang, X.; Beavers, L.S.; Wang, X.; Wang, J.; Gonzalez Valcarcel, I.C.; Ma, T. GPR142 Controls Tryptophan-Induced Insulin and Incretin Hormone Secretion to Improve Glucose Metabolism. PLoS ONE 2016, 11, e0157298.
- Chouchani, E.T.; Kazak, L.; Spiegelman, B.M. New Advances in Adaptive Thermogenesis: UCP1 and Beyond. Cell Metab. 2019, 29, 27–37.
- Nicholls, D.G. The hunt for the molecular mechanism of brown fat thermogenesis. Biochimie 2017, 134, 9–18.
- Wang, Z.; Wang, Q.A.; Liu, Y.; Jiang, L. Energy metabolism in brown adipose tissue. FEBS J. 2021, 288, 3647–3662.
- Yoneshiro, T.; Wang, Q.; Tajima, K.; Matsushita, M.; Maki, H.; Igarashi, K.; Dai, Z.; White, P.J.; McGarrah, R.W.; Ilkayeva, O.R.; et al. BCAA catabolism in brown fat controls energy homeostasis through SLC25A44. Nature 2019, 572, 614–619.
