You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 1 by Juan Han and Version 2 by Rita Xu.
Economic profit-driven food adulteration has become widespread in the dairy industry. One of the most common forms of dairy adulteration is the substitution of low-priced milk for high-priced milk. This has prompted regulatory authorities to focus on various means of authenticity testing. So far, many methods have been developed. Since milk adulteration has been upgraded, which has forced the testing methods to meet the needs of detection, which include DNA-based PCR methods. PCR and PCR-derived methods exhibit multiple advantages for authenticity testing, such as high stability, fast speed, and high efficiency, which meet the needs of modern testing.
- PCR
- adulteration identification method
- dairy products
1. Introduction
Food fraud has become a worldwide issue [1], resulting in huge economic losses and health risks [2]. Dairy products, which are made of essential nutrients, are the ideal food for the majority of humans, and thus, a huge consumer market and an adulteration epicenter have been formed [3][4][3,4]. With the enhanced seriousness of dairy product adulteration, the requirement for cheap, sensitive, and efficient methods for adulteration tests has been significantly increased. The adulteration of dairy induction comes in diverse forms such as the abuse of additives, the addition of milk extracts, the dilution with water, and the mixture of low-priced milk [5]. One common form of dairy product adulteration is the addition of low-priced milk to high-priced milk. Gonçalves et al. (2012) identified the source of commercial dairy products in Porto, and the results showed that 12.5% (12/96) of samples failed to confirm the description contents [6]. Similarly, Di Pinto et al. (2017) tested the authenticity of 80 goat cheeses from the Italian market and found that 80% were adulterated with cow/sheep milk [7]. Furthermore, Zhang et al. (2022) detected the source of 46 commercial minor species’ milk from the Chinese market, in which 15 samples were species false labeling [8]. Such adulteration can put the health and even the life of the consumer at risk, since any milk protein of unknown origin, especially cow milk protein, is a potential allergen [9][10][11][9,10,11]. Based on this, adulteration identification aimed at species-specific traits and quantitative estimation of adulteration levels are attractive to the supervisory authority.
In the past decades, numerous species-specific adulteration identification methods were developed based on the characteristic substances such as proteins, lipid acids, sugars, and DNA in dairy products. Protein-based identification tools, such as isoelectric-focusing, chromatography, and immunochemical technologies, exhibit advantages such as being fast, efficient, and affordable, and they are considered to be appropriate for species identification in dairy products [12][13][14][12,13,14]. However, instability of the protein structure tends to result in identification bias, limiting the application of protein-based identification in processed dairy products [12][13][12,13]. Although the mass spectrometry (MS) method allows accurate species identification in processed dairy products using marker peptides, the requirements for professional equipment and personnel significantly restrict its wide application [13][14][13,14]. In addition, many spectroscopic methods targeting at a feature group of nutrients (mainly including lipid acid, protein, and carbohydrate) have been developed for dairy product adulteration identification, such as mid-infrared (MIR), near-infrared (NIR), front-face fluorescence spectroscopy (FFFS), Fourier-transform infrared (FT-IR), and nuclear magnetic resonance (NMR) [13][14][13,14]. Although spectroscopic methods are more suitable for daily monitoring than other methods, the exorbitant equipment costs restrict their application range [8]. With the development of polymerase chain reaction (PCR), many techniques for evaluating authenticity by estimating the residual DNA sequence in dairy products have emerged and are extensively used in the dairy industry [15][16][15,16].
The thermal stability of DNA is higher than that of other nutrients in dairy products, and more importantly, the detection of DNA is independent from immune responses [12][13][12,13]. These advantages have encouraged the development of PCR-based assays, and a variety of PCR assay forms have been derived, which play a key role in dairy industry adulteration identification (Figure 1). The quality of the DNA extraction is closely related to the successful implementation of these PCR-based methods [17]. The composition of dairy products greatly affects the extraction of DNA, and accordingly affects the subsequent amplification and testing [18]. Therefore, the development of appropriate DNA extraction methods is essential for the promotion and improvement of PCR-based methods.
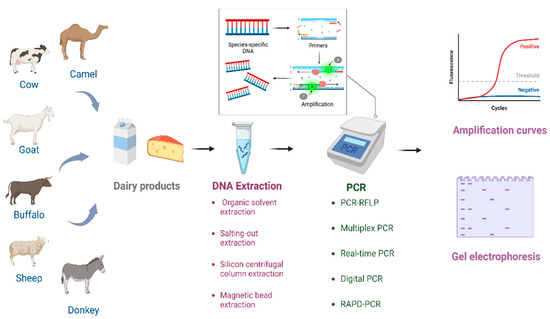
Figure 1. The process of identifying the authenticity of dairy products using PCR technology.
2. PCR-Based Dairy Product Authenticity Testing
PCR is widely used for animal origin identification in the dairy industry. The different species-specific DNA targets can be amplified by PCR, separated by agarose gel according to the fragment size, and visualized by DNA dye [19][20][38,39]. PCR has multiple advantages; for example, it is simple, less time-consuming, and it does not rely on expensive instruments [21][40]. DNA sequences with high copy numbers and high species specificity are used as targets for PCR assays. Mitochondrial DNAs with high inter-species specificity and high intra-species conservation are usually used as targets for PCR assays. Meanwhile, some nuclear DNA can also be used as targets for PCR assays. Recently, more and more derivative methods have been developed to improve PCR techniques for dairy product adulteration identification [12][13][14][12,13,14] such as PCR-restriction fragment length polymorphism (PCR-RFLP), multiplex PCR, real-time PCR, and digital PCR (dPCR) based on known DNA target sequences as well as random amplified polymorphic DNA-PCR (RAPD-PCR) developed based on unknown target sequences.
2.1. PCR-RFLP
RFLP-PCR is a simple, fast, and inexpensive method. It starts with sequence amplification by PCR, followed by enzymatic digestion with a restriction endonuclease, conducts separation by electrophoresis, and ends up with visualization by a UV gel imager. This method was first proposed by Plath et al. in 1997 to identify cow milk from sheep milk, goat milk, and cheese with polyacrylamide gel electrophoresis followed by PCR-RFLP [22][41]. Then, Abdel-Rahman et al. (2007) used PCR-RFLP and agarose gel to identify buffalo milk, cow milk, and sheep milk with the cytochrome b (cyt b)gene as the DNA target [23][42]. With the aid of computational biology, Lanzilao et al. (2005) set up an enzyme mixture (HaeIII, TaqI, and MwoI) acting on the cyt b gene to identify the milk sources from cow, sheep, goats, and buffaloes [24][43]. Then, Vafin et al. (2022) mixed XbaI and PsiI to digest the κ-casein gene and generate the RFLP profiles, enabling the identification of the milk sources (sheep, goat, cow, and buffalo) of dairy products [25][44].
2.2. Multiplex PCR
The multiplex PCR (mPCR) technique has been developed to increase sensitivity and efficiency. The mPCR can amplify multiple targets in a single PCR reaction to identify the multi-sourced dairy products with the advantages of high efficiency, high throughput, and inexpensiveness. This method was first developed by Bottero et al. (2003) to simultaneously detect multiple animal sources in dairy products with a detection limit of 0.5%. Multiplex PCR was used for the detection of laboratory-made cheeses (from cow milk, goat milk, and sheep milk) and commercial Italian cheeses with mitochondrial 12S and 16S rRNA genes as the targets [26][45]. Tsirigoti et al. (2020) designed a primer mixture with two cyt b genes based on cow and sheep and one DNA sequence based on the goat D-loop range to identify the milk sources, exhibiting a detection limit of 0.1% [27][46]. With the advancements in molecular biology technology, many quantitative PCR-based multiplex PCRs have been developed for the detection of dairy products.
2.3. Real-Time PCR
Real-time PCR is also known as quantitative real-time PCR, and it can monitor the amplification products in each PCR cycle using fluorescent reporter molecules. This real-time PCR method is gradually replacing traditional PCR due to its excellent sensitivity, specificity, and high consistency, as well as qualitative and quantitative properties. Recently, the qPCR has been classified into two types based on the sequence detection system. In one type, DNA is detected by double-stranded DNA-embedded dye represented by SYBR Green, and in the other type, DNA is detected by fluorophore-labelled oligonucleotides (represented by TaqMan). These two types of qPCR have their own advantages and different application scenarios. The qPCR using DNA dye has the advantage of being economical and rapid, but it has its own disadvantages such as low specificity and the requirement for a lysis curve analysis after amplification. In contrast, the qPCR using fluorophore-labeled oligonucleotides has the advantages of high specificity and the ability to set multiple detection targets, thus expanding its application to dairy product adulteration detection.
The qPCR with SYBR Green was first used by Hebert et al. (2003) to quantify the ratio of cow milk to buffalo milk in mozzarella cheese with cytochrome oxidase subunit 1 (coI) as the target gene [28][47]. Lopez-Calleja et al. (2007) first reported that qPCR with TaqMan probes could identify goat milk or cow milk from sheep milk by amplifying the 12S rRNA sequence, with a linear dynamic range of 0.6–10% [29][48]. The target DNA and genes used in qPCR are usually the mitochondrial 12S rRNA gene and cyt b gene. Numerous qPCR-based derivative methods, such as multiplex primer design and high-resolution melting (HRM) analysis, have been developed to extend their application scope.
Cottenet et al. (2011) first designed two probes with cyt b genes from cow and buffalo as targets, making the detection of two milk sources possible [30][49]. Later, duplex PCR based on 16S rRNA and D-LOOP genes was used to detect adulterated milk sources (camel, horse, and goat) with a detection limit of 0.1% in raw milk and 0.5% in ultra-high-temperature (UHT)-sterilized milk [31][50]. The triple qPCR is developed to simultaneously detect the target 12S rRNA gene of the species to be detected and the endogenous control by TaqMan probes, based on which the simultaneous milk source detection of cow and mare, cow and goat, sheep and goat, and camel and cow has been successfully implemented [32][33][34][35][51,52,53,54]. Agrimonti et al. (2015) developed a quadruplex PCR targeting the cyt b gene and 12S rRNA to identify and quantify milk sources from cow, sheep, goat, and buffalo with a detection limit of 0.1% [36][55].
As a highly efficient and robust PCR technique, high-resolution melting (HRM) analysis is not limited by mutation sites or types, and it is used to analyze mutations, single nucleotide polymorphisms (SNPs), methylation, and mapping of samples without requiring sequence-specific probes [37][56]. Like many fluorescent PCR techniques, HRM inserts a specific dye into the double-stranded DNA to detect the samples by real-time monitoring of the binding of the fluorescent double-stranded DNA dye to the PCR amplification product and recording a high-resolution melting curve. Compared with conventional PCR methods, HRM analysis is performed in the same tube as the PCR amplification step, and thus HRM requires no purification or separation of amplicons [38][57]. This makes HRM analysis faster, less laborious, and more suitable for high-throughput assays than other methods, thus increasing its applicability for food identification, especially for dairy product identification [39][58]. Ganopoulos et al. (2013) first reported that HRM analysis could be used to detect and quantify the cow milk in commercial samples of Feta cheeses (protected designation of origin, PDO), samples of sheep-goat cheeses (non-PDO), and samples of goat cheeses. In their report, the D-loop sequences of cattle and sheep and the tRNA-Leu gene of goats were used as targets to identify the authenticity of the label with a detection limit of 0.1% [40][59].
2.4. Digital PCR (dPCR)
To further reduce the detection limit, digital PCR (dPCR) has also been used to test the authenticity of dairy products. dPCR is an absolutely quantitative PCR, and it starts with the limited dilution of PCR reactants, followed by PCR amplification in different reaction chambers, and ends with qualification of initial copy number or concentration of target molecules based on the Poisson distribution principle and the number and proportion of positive microdroplets [41][60]. This method was developed in the late 1980s [42][61], and it was first named digital PCR in 1999 [43][62]. Compared to qPCR, dPCR has the following advantages: sensitivity up to the single-molecule level, detection limit as low as 0.001%, ability to absolutely quantify initial levels of target molecules without requiring standard curves, and independence from Ct value, PCR amplification efficiency, and PCR inhibitor effects [44][63]. In the past, dPCR was widely used in the dairy testing industry to detect pathogenic microbial contamination in dairy products, and at present, it is used to identify milk sources [45][46][47][48][64,65,66,67]. Cutarelli et al. (2021) used the droplet digital PCR (ddPCR) with cyt b gene as the target to identify the cow and buffalo milk in mozzarella cheese [45][64].
3. Factors Affecting PCR-Based Dairy Product Authenticity Testing
DNA is a stable molecular marker for food authenticity testing. Food fraud detection methods based on PCR have been emerging in the past decades. However, many factors upstream of PCR detection limit its application in dairy product authenticity testing.
3.1. Disruption of DNA Integrity by Food Processing
DNA integrity affects downstream PCR amplification, which in turn affects the accuracy of the assay. Most dairy products, such as butter, cream, cheese, and UHT-sterilized milk, are heat-processed to extend their shelf life, which can result in DNA degradation [49][78]. During the preparation of milk powder, denatured proteins can bind to DNA, thus reducing the purity of the extracted DNA [50][79]. In addition to the effect of heating, the ripening process of cheese can also reduce the integrity of DNA, thereby affecting DNA amplification detection [51][80]. The DNA of the microorganisms that play a role in cheese ripening also reduces the purity of the milk DNA, hence influencing the PCR detection [52][81].
3.2. PCR Inhibitors
Many components that inhibit the PCR reaction are found in dairy products. For example, the fat and protease in cheese and butter, and even certain metal ions, such as calcium ions in milk, are PCR inhibitors [48][53][54][55][67,82,83,84]. Furthermore, PCR inhibitors, such as EDTA, polysaccharides, and other organic solvents, may be introduced during the DNA extraction process [56][85].
