Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Jessie Wu and Version 1 by Alexander Zhgun.
L-asparaginases (L-ASNases) of microbial origin are the mainstay of blood cancer treatment. Numerous attempts have been performed for genetic improvement of the main properties of these enzymes. The substrate-binding Ser residue is highly conserved in L-ASNases regardless of their origin or type. However, the residues adjacent to the substrate-binding Ser differ between mesophilic and thermophilic L-ASNases.
- extremophilic enzyme
- hyperthermophile
- L-asparaginase
- substrate-binding
- mutagenesis
- enzymatic activity
1. Introduction
Among currently used enzymes, L-asparaginase (L-ASNase) remains one of the most important used in medicine, biosensors, and food industry. L-ASNase (EC 3.5.1.1; L-asparagine amidohydrolase) catalyzes the hydrolysis of L-asparagine (L-Asn) to L-aspartic acid and ammonia [1]. It is the first enzyme with antitumor activity to be used in clinical practice after its approval in 1978 [2,3][2][3]. Currently, L-ASNase continues to be the mainstay for the treatment of pediatric acute lymphoblastic leukemia and is also used to treat other related blood cancers worldwide [4,5][4][5].
L-ASNase therapy is based on L-Asn starvation of susceptible tumor cells that are unable to synthesize their own L-Asn. By hydrolyzing extracellular L-Asn, L-ASNase leads to the death of lymphoblastic cells by apoptosis [6,7][6][7].
Relatively recently, the biotechnological area of L-ASNase application has expanded. In 2002, it was discovered that high contents of acrylamide are formed between reducing sugars and L-asparagine during high-temperature (over 120 °C) processing of starchy foods under low humidity in a non-enzymatic Maillard reaction [8].
By hydrolyzing L-Asn, L-ASNase prevents the formation of carcinogenic acrylamide in the Maillard reaction [9]. As aspartic acid cannot participate in the reaction, L-ASNase treatment helps to reduce the formation of acrylamide in commercially fried foods. The method is safe and effective: L-ASNase treatment leads to a decrease in the content of acrylamide up to 99%, and further heating deactivates the enzyme without affecting the properties of the final product [10,11][10][11].
Another field of the enzyme application is the development of biosensors. Biosensor systems based on L-ASNase allow to detect the level of L-Asn in medicine and in the food industry [12,13][12][13].
Thus, L-ASNase is an enzyme that is widely used in biotechnology under various operating conditions, in particular, in a wide range of temperatures. Most L-ASNases exhibit optimal activity at or near mesophilic temperatures (approximately 30–40 °C) and under mild operating conditions [2]. Obviously, it is difficult for enzymes of mesophilic origin to cover such a wide range of conditions required for their successful application in biotechnology, especially in the food industry. Extremophiles, in particular, thermophiles, which have been reported to produce L-ASNases with unique properties, can expand the number of biotechnologically available L-ASNases [9,14,15,16,17][9][14][15][16][17]. According to experimental data, thermophilic L-ASNases (thermo-ASNases) can not only occupy a vacant niche of high-temperature food technologies but also compete with mesophilic enzymes in biomedicine [9,15,16,17][9][15][16][17].
Due to their superior performance, investigation of thermo-ASNases is of particular interest. Elucidation of the molecular mechanisms that determine the relationship between the activity, stability, and flexibility of thermo-ASNases is critical for both fundamental and applied research in view of their biotechnological significance.
Previously, wresearchers have characterized a new promising hyperthermophilic L-ASNase from the archaea Thermococcus sibiricus (TsA) [16]. The enzyme is optimally active at 90 °C, stable, and exhibits high specific activity and strong cytotoxic activity toward cancer cells.
Various protein engineering approaches are used in an attempt to improve the properties of known L-ASNases. Based on the traditional directed evolution method, Kotzia and Labrou engineered mutants of mesophilic L-ASNases from Erwinia carotovora and Erwinia chrysanthemi [18]. A thermostable mutant Asp133Val was obtained from a library of enzyme variants. This powerful approach is highly dependent on the quality and quantity of the generated libraries, and a target biobetter form is hard to obtain [19,20][19][20]. Enzyme computational engineering approaches can reduce search screening. Offman et al. successfully constructed a proteolysis resistant mutant of Escherichia coli L-ASNase with improved activity by adapting a genetic algorithm of protein modeling in combination with molecular dynamics flexibility studies [21]. The sequence-based approach is a useful technique for engineering desired forms of proteins, in particular with unsolved structures, based on the analysis of well-characterized homologous enzymes. By employing multiple sequences alignment in tandem with homologous modeling, mutant form of Bacillus subtilis L-ASNase with improved thermostability was obtained [22]. Based on sequence alignment and structure superposition of thermophilic and mesophilic L-ASNases, Li et al. identified two residues that affect their thermostability [23].
2. Enhancing the Catalytic Activity of Thermo-Asparaginase from Thermococcus sibiricus
Mutations usually have a variety of abilities to fine-tune the functions of enzymes [34,35][24][25]. No general guidelines to enhance the activity of the promising wild-type enzyme have been established. For thermo-ASNase TsA, the approach based on the simultaneous mesophilic-like substitutions of residues adjacent to the substrate-binding Ser in the highly conserved DST triad resulted in a 2-fold increase in activity.
Homology models revealed that a single mutation—either Asp (thermo-ASNase) → Gly (meso-ASNase EcAII) or Thr (thermo-ASNase) → Gln (meso-ASNase EcAII)—impairs proper substrate binding. Nevertheless, restoring the entire mesophilic-like triad GSQ in TsA after double mutation improved its activity.
In EcAII, the downstream residue Gln59 assists the Ser58 residue in substrate binding. In thermo-ASNases, the upstream Asp residue adjacent to the substrate-binding Ser is supposed to play the same role. The puzzle is that, in terms of function, the DST triad is “inverse” in thermo-ASNases compared to mesophilic GSQ relative the substrate-binding Ser. Thus, both previously used mutational strategies in this region to replace single Asp or Thr residue with their directly corresponding residues of EcAII are not entirely correct from the point of increasing activity.
In thermo-ASNase PfA, the T53Q mutant is reported to have an increased substrate affinity of 8.3 mM compared to 12.1 mM for the wild-type enzyme. At the same time, the substitution of conservative Thr53 correlates with a more than 2-fold loss in catalytic efficiency [15]. The combination of three adjacent DSQ residues involved in substrate binding and stabilization can lead to an increase in docking strength in thermo-ASNases. At high docking strength, the docking lifetime is longer than the time required for catalysis [36][26].
In turn, in thermo-ASNases Pyrococcus yayanosii (PyA) and Thermococcus gammatolerans (TgA), the Asp → Gly replacement with the resulting “mixed” triad residues GST impaired substrate binding, increased flexibility around the binding Ser residue due to the deficiency of polar contacts [23]. Decreased binding function of the Ser residue caused dramatic loss of PyA and TgA activity (Table 1).
Table 1.
Effect of residues adjacent to the substrate-binding Ser on the main characteristics of thermophilic L-ASNases.
| Combination of Residues Adjacent to the Catalytic Active Ser * | Source of L-ASNase | Abbreviation | Topt | Km Value, mM | Specific Activity, U/mg | References | |||
|---|---|---|---|---|---|---|---|---|---|
| mesophilic G | S58 | Q | E. coli | EcAIIwt | 37 | 0.018 | 235 | [23] | |
| D | S52 | T | P. furiosus | PfAwt | 80–85 | 12.1 | 550 | [15,37] | [15][27] |
| D | S52 | Q | P. furiosus | PfAmut | 80 | 8.3 | ND | [15] | |
| D | S52 | T | P. yayanosii | PyAwt | 95 | 6.5 | 1483 | [23] | |
| G | S52 | T | P. yayanosii | PyAmut | 65 | 3.1 | 341 | [23] | |
| D | S53 | T | T. gammatolerans | TgAwt | 85 | 8.3 | 5381 | [23] | |
| G | S53 | T | T. gammatolerans | TgAmut | 60 | 6.8 | 962 | [23] | |
| D | S55 | T | T. sibiricus | TsAwt | 90 | 3.0 | 2066.1 | [16], this study | [16], this research |
| G | S55 | Q | T. sibiricus | TsAmut | 90 | 6.0 | 5037.7 | This study | This research |
* GSQ—mesophilic, DST—thermophilic, other combinations are “mixed”.
Although single mesophilic-like substitutions of residues adjacent to the substrate-binding Ser—Thr→Gln and Asp→Gly reduced the catalytic efficiency or activity of thermo-ASNases, the simultaneous introduction of their combination in TsA increased the specific activity of the enzyme (Table 1).
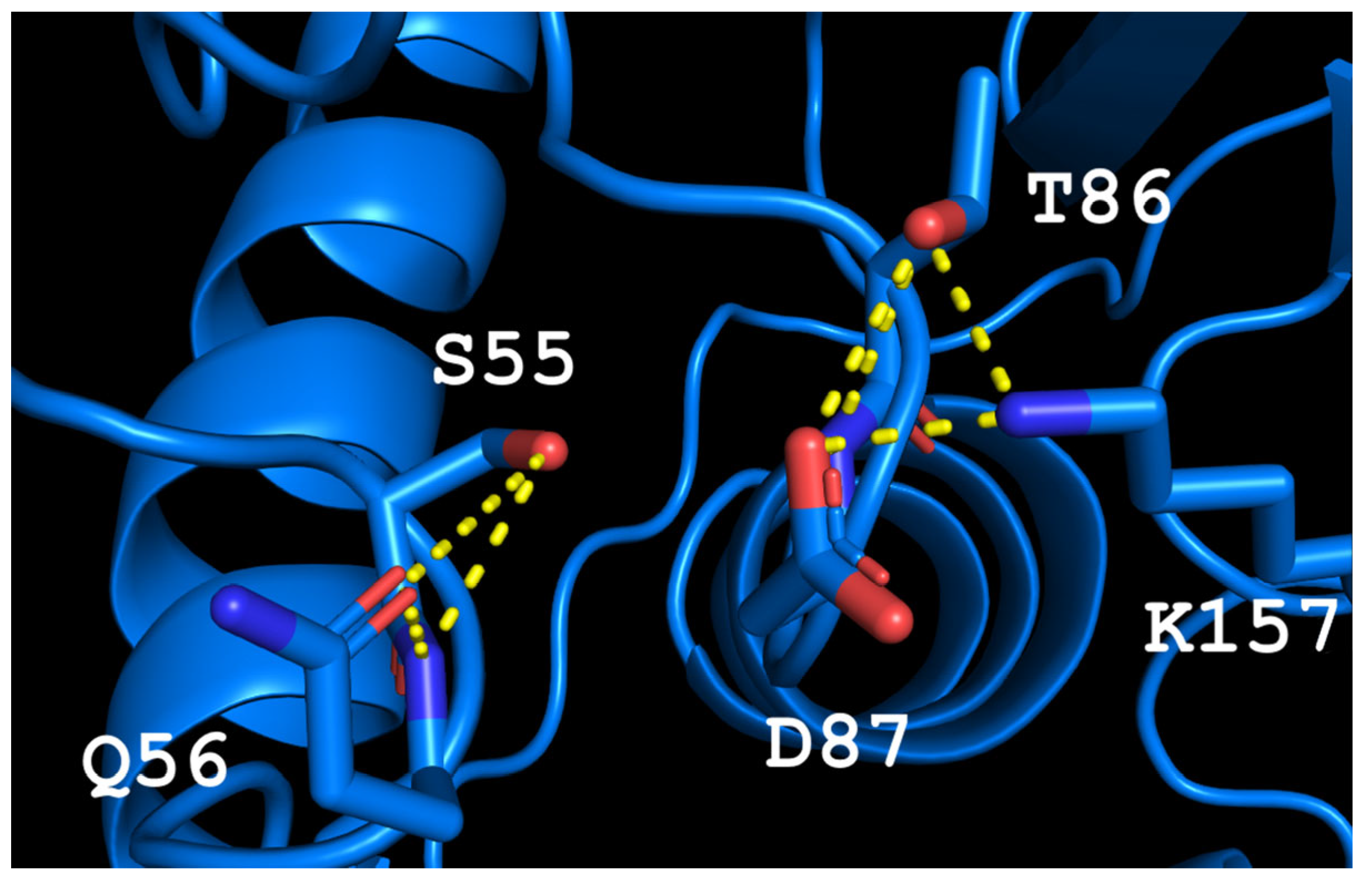
The predicted model of TsA showed that the structure of the active site is rigid, stabilized by a network of H-bonds. The T56Q mutation disrupts this network, increasing the flexibility of the active site, but the appearance of a bulky glutamine side chain can affect the interaction of the substrate in the active site. This observation leads to the mutation of the second residue to increase the available volume to accommodate the glutamine side chain. Thus, the second mutation D54G was chosen. The possible conformation of residues in the active site of the double mutant is shown in Figure 1. The resulting GSQ triad, fine-tuned to substrate binding in mesophilic EcAII, provided proper “clamping” and stabilizing of the substrate during the catalytic process in TsA thermo-ASNase. The increased flexibility of the structure around the active site may promote reorientation, substrate binding and increase the specific activity.
 Analysis of the mutant specific activity revealed that the conjoint substitution D54G/T56Q led to a significant increase in activity toward L-Asn—from 2066.1 U/mg for TsAwt to 5037.7 U/mg for the double mutant. In addition, a slight decrease in glutaminase activity was observed: from 7% for TsAwt to 5% for the mutant D54G/T56Q. Consistent with ouresearchers' data, in studies of EcAII, it was shown that Gly57 and Gln59 residues can modulate the substrate specificity of L-ASNase [23,26,27][23][28][29].
By analyzing the effect of temperature on the activity and stability of the double mutant, it was revealed that there was no shift in the optimum temperature, while the sensitivity to temperature changes and time-dependent loss of absolute enzyme activity increased.
The effect of a single mutation corresponding to D54G of TsA—D51G of the thermo-ASNase PyA and D52G of the thermo-ASNase TgA was studied by Li et al. [23]. The mutated enzymes PyA D51G and TgA D52G displayed a lower optimum temperature: the Topt values were 30 °C and 25 °C lower than those of the wild-type enzymes, respectively (Table 31) [23].
In reverse experiments using representative mesophilic L-ASNases from E. coli EcAII and Bacillus subtilis BsAII, site-directed mutagenesis was carried out, where residues G57 of EcAII and G107 of BsAII were replaced with the corresponding residue D54 of TsA. In this restudyearch, EcAII-G57D showed higher Topt than wild-type EcAII [23]. In contrast, for BsAII-G107D, no shift in temperature optimum was found.
No shift in the optimum temperature was reported after the substitution of Thr53 in PfA corresponding to Thr56 of TsA [15].
Li et al. have shown that the replacement of the corresponding residue of TsA D54G—PyA D51G and TgA D52G—led to a decrease in the thermostability of thermo-ANSases, regardless of their source [23], which is consistent with ouresearchers' experimental data. Conversely, the substitution G→D improved thermostability for both mesophilic L-ASNases—EcAII and BsAII [22,23][22][23].
The overall results confirm that the amino acid residue in the corresponding position—D54 of TsA, D52 of TgA, D51 of PyA, G57 of EcAII, and G107 of BsAII—is one of the key residues responsible for the thermostability of L-ASNases, but this residue does not necessarily affect L-ASNase optimum temperature.
Analysis of cytotoxic activity in vitro revealed that the double mutant D54G/T56Q was more active against cancer cell lines than TsAwt. The IC90 values were 2.8-fold to 7.4-fold lower for the mutant than for the wild-type enzyme. A drastic increase in cytotoxic activity was also previously reported for the PfA mutant T53Q [15].
The triad of L-ASNases studied in this woreseark ch is actually a special case illustrating a key substitution between thermophilic and mesophilic enzymes. Known thermo-ASNases, including TsA, avoid the uncharged polar residue Gln [14]. Indeed, EcAII contains 13 Gln residues, while only 3 glutamines are present in TsA. In general, a decreased content of Gln residues in enzymes of thermophilic origin is a common feature. At higher temperatures, the frequency of spontaneous chemical modifications, such as deamidation, increases multiple folds [38][30]. The reaction rate of deamidation increases 350-fold at 100 °C compared to 37 °C. The deamidation mechanism is known for two residues—Asn and Gln [39][31]. The absence of a unique unstable Gln residue in thermo-ASNases, which promotes substrate binding in mesophilic GSQ triad, is supposed to minimize the possibility of chemical modifications and prevent blocking of protein functioning.
A small Thr residue of thermo-ASNases involved in multiple interactions in this region contributes to protein packing density and structural rigidity at high temperatures.
If the environment is cooler, resistance to spontaneous modification and thermal stability are not so important. The fitness of mesophiles is determined by enzyme activity, which is affected by the flexibility of the protein structure. According to structure modeling, the Gln residue in the mesohilic EcAII GSQ triad is preferable for increasing conformational flexibility of L-ASNase active site.
Taking into consideration, that susceptible to destruction and modification at high temperatures amino acid residues can also present in thermophilic and even hyperthermophilic enzymes if they are involved in specific stabilizing interactions or bring a special function and/or are inaccessible to the solvent [39][31], the T56Q mutation was performed. The thermolabile Gln residue was introduced relying on conformational environment and mobile lid cover the active site as protection factors. Neighboring Asp was replaced by Gly to keep the proper active site architecture favoring catalysis.
In this restudyearch, the conjoint substitution D54G/T56Q increased flexibility around the active site of TsA, facilitating conformational changes upon substrate binding and resulting in increased activity. As expected, the mutant displayed slightly lower heat stability. Enhanced rigidity of thermophilic enzymes correlates with increased thermal stability and vice versa [39,40][31][32].
An approach based on pairwise sequence comparison of thermophilic/mesophilic L-ASNases and substitution of highly conserved amino acid “special residues” by beneficial non-conflicting residues of mesophilic EcAII improved the catalytic activity of TsA thermo-ASNase. In TsA, the GSQ triad, fine-tuned for substrate binding in mesophilic EcAII, increased activity more than 2-fold at 37 °C and 90 °C. D54G/T56Q double mutant with increased activity at 90 °C can be efficiently used in the high-temperature food industry. The increased activity at 37 °C and cytotoxicity of TsA-D54G/T56Q make it possible to compete with other L-ASNases in biomedicine.
Analysis of the mutant specific activity revealed that the conjoint substitution D54G/T56Q led to a significant increase in activity toward L-Asn—from 2066.1 U/mg for TsAwt to 5037.7 U/mg for the double mutant. In addition, a slight decrease in glutaminase activity was observed: from 7% for TsAwt to 5% for the mutant D54G/T56Q. Consistent with ouresearchers' data, in studies of EcAII, it was shown that Gly57 and Gln59 residues can modulate the substrate specificity of L-ASNase [23,26,27][23][28][29].
By analyzing the effect of temperature on the activity and stability of the double mutant, it was revealed that there was no shift in the optimum temperature, while the sensitivity to temperature changes and time-dependent loss of absolute enzyme activity increased.
The effect of a single mutation corresponding to D54G of TsA—D51G of the thermo-ASNase PyA and D52G of the thermo-ASNase TgA was studied by Li et al. [23]. The mutated enzymes PyA D51G and TgA D52G displayed a lower optimum temperature: the Topt values were 30 °C and 25 °C lower than those of the wild-type enzymes, respectively (Table 31) [23].
In reverse experiments using representative mesophilic L-ASNases from E. coli EcAII and Bacillus subtilis BsAII, site-directed mutagenesis was carried out, where residues G57 of EcAII and G107 of BsAII were replaced with the corresponding residue D54 of TsA. In this restudyearch, EcAII-G57D showed higher Topt than wild-type EcAII [23]. In contrast, for BsAII-G107D, no shift in temperature optimum was found.
No shift in the optimum temperature was reported after the substitution of Thr53 in PfA corresponding to Thr56 of TsA [15].
Li et al. have shown that the replacement of the corresponding residue of TsA D54G—PyA D51G and TgA D52G—led to a decrease in the thermostability of thermo-ANSases, regardless of their source [23], which is consistent with ouresearchers' experimental data. Conversely, the substitution G→D improved thermostability for both mesophilic L-ASNases—EcAII and BsAII [22,23][22][23].
The overall results confirm that the amino acid residue in the corresponding position—D54 of TsA, D52 of TgA, D51 of PyA, G57 of EcAII, and G107 of BsAII—is one of the key residues responsible for the thermostability of L-ASNases, but this residue does not necessarily affect L-ASNase optimum temperature.
Analysis of cytotoxic activity in vitro revealed that the double mutant D54G/T56Q was more active against cancer cell lines than TsAwt. The IC90 values were 2.8-fold to 7.4-fold lower for the mutant than for the wild-type enzyme. A drastic increase in cytotoxic activity was also previously reported for the PfA mutant T53Q [15].
The triad of L-ASNases studied in this woreseark ch is actually a special case illustrating a key substitution between thermophilic and mesophilic enzymes. Known thermo-ASNases, including TsA, avoid the uncharged polar residue Gln [14]. Indeed, EcAII contains 13 Gln residues, while only 3 glutamines are present in TsA. In general, a decreased content of Gln residues in enzymes of thermophilic origin is a common feature. At higher temperatures, the frequency of spontaneous chemical modifications, such as deamidation, increases multiple folds [38][30]. The reaction rate of deamidation increases 350-fold at 100 °C compared to 37 °C. The deamidation mechanism is known for two residues—Asn and Gln [39][31]. The absence of a unique unstable Gln residue in thermo-ASNases, which promotes substrate binding in mesophilic GSQ triad, is supposed to minimize the possibility of chemical modifications and prevent blocking of protein functioning.
A small Thr residue of thermo-ASNases involved in multiple interactions in this region contributes to protein packing density and structural rigidity at high temperatures.
If the environment is cooler, resistance to spontaneous modification and thermal stability are not so important. The fitness of mesophiles is determined by enzyme activity, which is affected by the flexibility of the protein structure. According to structure modeling, the Gln residue in the mesohilic EcAII GSQ triad is preferable for increasing conformational flexibility of L-ASNase active site.
Taking into consideration, that susceptible to destruction and modification at high temperatures amino acid residues can also present in thermophilic and even hyperthermophilic enzymes if they are involved in specific stabilizing interactions or bring a special function and/or are inaccessible to the solvent [39][31], the T56Q mutation was performed. The thermolabile Gln residue was introduced relying on conformational environment and mobile lid cover the active site as protection factors. Neighboring Asp was replaced by Gly to keep the proper active site architecture favoring catalysis.
In this restudyearch, the conjoint substitution D54G/T56Q increased flexibility around the active site of TsA, facilitating conformational changes upon substrate binding and resulting in increased activity. As expected, the mutant displayed slightly lower heat stability. Enhanced rigidity of thermophilic enzymes correlates with increased thermal stability and vice versa [39,40][31][32].
An approach based on pairwise sequence comparison of thermophilic/mesophilic L-ASNases and substitution of highly conserved amino acid “special residues” by beneficial non-conflicting residues of mesophilic EcAII improved the catalytic activity of TsA thermo-ASNase. In TsA, the GSQ triad, fine-tuned for substrate binding in mesophilic EcAII, increased activity more than 2-fold at 37 °C and 90 °C. D54G/T56Q double mutant with increased activity at 90 °C can be efficiently used in the high-temperature food industry. The increased activity at 37 °C and cytotoxicity of TsA-D54G/T56Q make it possible to compete with other L-ASNases in biomedicine.

Figure 1.
The model of the TsA-T56Q/D54G active site. Yellow dash lines—H-bonds.
References
- Lopes, A.M.; de Oliveira-Nascimento, L.; Ribeiro, A.; Tairum, C.A.; Breyer, C.A.; de Oliveira, M.A.; Monteiro, G.; de Souza-Motta, C.M.; Magalhães, P.d.O.; Avendaño, J.G.F.; et al. Therapeutic l-asparaginase: Upstream, downstream and beyond. Crit. Rev. Biotechnol. 2017, 37, 82–99.
- Lubkowski, J.; Wlodawer, A. Structural and biochemical properties of L-asparaginase. FEBS J. 2021, 288, 4183–4209.
- Hill, J.M.; Roberts, J.; Loeb, E.; Khan, A.; Maclellan, A.; Hill, R.W. L-Asparaginase Therapy for Leukemia and Other Malignant Neoplasms: Remission in Human Leukemia. JAMA J. Am. Med. Assoc. 1967, 202, 882–888.
- Ghasemian, A.; Al-marzoqi, A.H.; Al-abodi, H.R.; Alghanimi, Y.K.; Kadhum, S.A.; Shokouhi Mostafavi, S.K.; Fattahi, A. Bacterial l-asparaginases for cancer therapy: Current knowledge and future perspectives. J. Cell. Physiol. 2019, 234, 19271–19279.
- Maggi, M.; Scotti, C. Enzymes in metabolic anticancer therapy. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2019; Volume 1148.
- Mahajan, R.V.; Kumar, V.; Rajendran, V.; Saran, S.; Ghosh, P.C.; Saxena, R.K. Purification and characterization of a novel and robust L-asparaginase having low-glutaminase activity from bacillus licheniformis: In vitro evaluation of anti-cancerous properties. PLoS ONE 2014, 9, e99037.
- Ali, U.; Naveed, M.; Ullah, A.; Ali, K.; Shah, S.A.; Fahad, S.; Mumtaz, A.S. L-asparaginase as a critical component to combat Acute Lymphoblastic Leukaemia (ALL): A novel approach to target ALL. Eur. J. Pharmacol. 2016, 771, 199–210.
- Stadler, R.H.; Blank, I.; Varga, N.; Robert, F.; Hau, J.; Guy, P.A.; Robert, M.C.; Riediker, S. Acrylamide from Maillard reaction products. Nature 2002, 419, 449–450.
- Sajed, M.; Naeem, S.U.; Rashid, N. L-Asparaginases from hyperthermophilic archaea and their applications. In Microbial Extremozymes; Academic Press: Cambridge, MA, USA, 2022; pp. 177–184.
- Gazi, S.; Göncüoğlu Taş, N.; Görgülü, A.; Gökmen, V. Effectiveness of asparaginase on reducing acrylamide formation in bakery products according to their dough type and properties. Food Chem. 2023, 402, 134224.
- Mohan Kumar, N.S.; Shimray, C.A.; Indrani, D.; Manonmani, H.K. Reduction of Acrylamide Formation in Sweet Bread with l-Asparaginase Treatment. Food Bioprocess Technol. 2014, 7, 741–748.
- Verma, N.; Kumar, K.; Kaur, G.; Anand, S. E. coli K-12 asparaginase-based asparagine biosensor for leukemia. Artif. Cells Blood Substit. Biotechnol. 2007, 35, 449–456.
- Kumar, K.; Kataria, M.; Verma, N. Plant asparaginase-based asparagine biosensor for leukemia. Artif. Cells Nanomed. Biotechnol. 2013, 41, 184–188.
- Dumina, M.; Zhgun, A. Thermo-L-Asparaginases: From the Role in the Viability of Thermophiles and Hyperthermophiles at High Temperatures to a Molecular Understanding of Their Thermoactivity and Thermostability. Int. J. Mol. Sci. 2023, 24, 2674.
- Bansal, S.; Srivastava, A.; Mukherjee, G.; Pandey, R.; Verma, A.K.; Mishra, P.; Kundu, B. Hyperthermophilic asparaginase mutants with enhanced substrate affinity and antineoplastic activity: Structural insights on their mechanism of action. FASEB J. 2012, 26, 1161–1171.
- Dumina, M.; Zhgun, A.; Pokrovskaya, M.; Aleksandrova, S.; Zhdanov, D.; Sokolov, N.; El’darov, M. A Novel L-Asparaginase from Hyperthermophilic Archaeon Thermococcus sibiricus: Heterologous Expression and Characterization for Biotechnology Application. Int. J. Mol. Sci. 2021, 22, 9894.
- Nadeem, M.S.; Khan, J.A.; Al-Ghamdi, M.A.; Khan, M.I.; Zeyadi, M.A. Studies on the recombinant production and anticancer activity of thermostable L-asparaginase I from Pyrococcus abyssi. Braz. J. Biol. 2022, 82, e244735.
- Kotzia, G.A.; Labrou, N.E. Engineering thermal stability of l-asparaginase by in vitro directed evolution. FEBS J. 2009, 276, 1750–1761.
- Sen, S.; Venkata Dasu, V.; Mandal, B. Developments in directed evolution for improving enzyme functions. Appl. Biochem. Biotechnol. 2007, 143, 212–223.
- Bilal, M.; Iqbal, H.M.N. Tailoring Multipurpose Biocatalysts via Protein Engineering Approaches: A Review. Catal. Lett. 2019, 149, 2204–2217.
- Offman, M.N.; Krol, M.; Patel, N.; Krishnan, S.; Liu, J.Z.; Saha, V.; Bates, P.A. Rational engineering of L-asparaginase reveals importance of dual activity for cancer cell toxicity. Blood 2011, 117, 1614–1621.
- Long, S.; Zhang, X.; Rao, Z.; Chen, K.; Xu, M.; Yang, T.; Yang, S. Amino acid residues adjacent to the catalytic cavity of tetramer l-asparaginase II contribute significantly to its catalytic efficiency and thermostability. Enzyme Microb. Technol. 2016, 82, 15–22.
- Li, X.; Zhang, X.; Xu, S.; Xu, M.; Yang, T.; Wang, L.; Zhang, H.; Fang, H.; Osire, T.; Rao, Z. Insight into the thermostability of thermophilic L-asparaginase and non-thermophilic L-asparaginase II through bioinformatics and structural analysis. Appl. Microbiol. Biotechnol. 2019, 103, 7055–7070.
- Pokrovskaya, M.V.; Pokrovsky, V.S.; Aleksandrova, S.S.; Sokolov, N.N.; Zhdanov, D.D. Molecular Analysis of L-Asparaginases for Clarification of the Mechanism of Action and Optimization of Pharmacological Functions. Pharmaceutics 2022, 14, 599.
- Li, Y.; Song, K.; Zhang, J.; Lu, S. A computational method to predict effects of residue mutations on the catalytic efficiency of hydrolases. Catalysts 2021, 11, 286.
- Dyla, M.; González Foutel, N.S.; Otzen, D.E.; Kjaergaard, M. The optimal docking strength for reversibly tethered kinases. Proc. Natl. Acad. Sci. USA 2022, 119, e2203098119.
- Kengen, S.W.M. Pyrococcus furiosus, 30 years on. Microb. Biotechnol. 2017, 10, 1441–1444.
- Borek, D.; Kozak, M.; Pei, J.; Jaskolski, M. Crystal structure of active site mutant of antileukemic L-asparaginase reveals conserved zinc-binding site. FEBS J. 2014, 281, 4097–4111.
- Sanches, M.; Barbosa, J.A.R.G.; De Oliveira, R.T.; Neto, J.A.; Polikarpov, I. Structural comparison of Escherichia coli L-asparaginase in two monoclinic space groups. Acta Crystallogr. Sect. D Biol. Crystallogr. 2003, 59, 416–422.
- Geiger, T.; Clarke, S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 1987, 262, 785–794.
- Vieille, C.; Zeikus, G.J. Hyperthermophilic Enzymes: Sources, Uses, and Molecular Mechanisms for Thermostability. Microbiol. Mol. Biol. Rev. 2001, 65, 1–43.
- Dumina, M.V.; Eldarov, M.A.; Zdanov, D.D.; Sokolov, N.N. L-Asparaginases of Extremophilic Microorganisms in Biomedicine. Biochem. Suppl. Ser. B Biomed. Chem. 2020, 14, 33.
More
