Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 3 by Rita Xu and Version 2 by Rita Xu.
Filamentous fungi are one of the most important producers of secondary metabolites. Some of them can have a toxic effect on the human body, leading to diseases. On the other hand, they are widely used as pharmaceutically significant drugs, such as antibiotics, statins, and immunosuppressants. A single fungus species in response to various signals can produce 100 or more secondary metabolites. Such signaling is possible due to the coordinated regulation of several dozen biosynthetic gene clusters (BGCs), which are mosaically localized in different regions of fungal chromosomes.
- secondary metabolites
- biosynthetic gene clusters (BGCs)
- filamentous fungi
1. Introduction
The production of secondary metabolites (SMs) is one of the most prominent biochemical attributes of filamentous fungi (or moldy fungi, or molds), and has stimulated extensive research on these microorganisms since the 1950s [1][2][3]. As a result of this, numerous compounds have been discovered, some of which are capable of harming human health, while others are able to heal people [4][5][6][7]. In parallel with the emergence of new knowledge about the effects of these low-molecular-weight compounds on the human body and the deciphering of their structures, investigations have been carried out that aim to study the mechanism of action at the cellular level [8]. For example, targets for the main classes of antibiotics have been identified, and the mechanisms for the emergence of resistance in microorganisms against these drugs have been established [9][10][11][12][13][14]. In the late 1980s, in light of the emergence of the era of genetic engineering, the molecular basis of the biosynthesis of secondary metabolites began to be studied [15]. In particular, the so-called biosynthetic gene clusters (BGCs) responsible for the biosynthesis of the corresponding SMs were discovered [16][17]. This knowledge made it possible to apply the strategy of reverse genetics, i.e., going from gene to trait/phenotype, and search for an appropriate product for “silent” or so-called “orphan” BGCs [18][19][20]. Currently, there are various techniques for such genome-mining of BGCs [15][21][22][23][24][25]. Emerging knowledge about biosynthetic gene clusters for the production of secondary metabolites, as well as a lot of difficulties associated with the “awakening” of silent BGCs, have led to the understanding, in numerous studies, of the existence of a complex regulatory system for them [26][27][28][29]. Such regulation operates in concert at several levels, starting with cluster-specific regulators, transcription factors whose genes cluster within a particular BGC and regulate the expression of the same BGC, ending with global regulators and chromatin-mediated regulation [30][31][32][33]. Due to the presence of such a system of regulation, there is a relationship between the production of SMs and the development of fungi [34]. The production of the corresponding SMs occurs at certain stages of the development of the fungus; for example, the synthesis of pigments occurs after the transition from the growth phase (trophophase) to the production phase (idiophase) [35]. On the other hand, most fungal BGCs are silent under normal physiological conditions and begin to work after receiving an appropriate environmental signal that affects the regulatory system [36][37][38]. Composite pleiotropic events accompanying the functioning of the fungal secondary metabolism are currently being studied using complex, including multi-omics, approaches [39][40][41].
The existing fundamental knowledge of the biosynthesis and regulation of SMs in filamentous fungi is extremely important, since, based on natural isolates, over the past 70–80 years, numerous industrial producers of pharmaceutically significant drugs, such as antibiotics, statins, and immunosuppressants, have been created [2][42][43][44]. Numerous works are also underway to create strains-producers of antitumor drugs that are synthesized in fungi [45]. Such industrial producers have been obtained as a result of the so-called classical strain improvement (CSI) methods associated with random mutagenesis and screening for the production of targeted SMs [46][47]. Modern knowledge about the organization of the regulatory machinery of secondary metabolism in the fungal cell makes it possible to understand the molecular basis of the direction of mutational selection, leading to high-yield production of the target secondary metabolite [48]. To achieve this, the original wild-type strains and improved producers are compared at the reference points of improvement programs [49]. Understanding the changes that have taken place is important for the development of future approaches to the targeted genetic engineering of high-yielding fungal producers of SMs [50][51].
2. Main Types of Fungal Secondary Metabolites
Fungi are one of the most evolutionarily adapted organisms, which has allowed them to occupy the majority of ecological niches suitable for existence on Earth over the past billion years [52][53]. According to existing estimates, global fungal diversity is about an order of magnitude greater than that of land plants [54][55]. One of the paramount assistants to such adaptive expansion was the ability to produce wide-variable low-molecular compounds, the so-called secondary metabolites, in response to changes in the state of both the organism itself and the environment [56][57][58]. These highly active molecules have begun to play a trigger function, and are selected as keys to the locks of various processes in the development of the organism itself, and its defense and/or attack against surrounding organisms and other species via within- and between-species interaction. More than 15,000 biologically active SMs are currently known to be produced by fungi (which is approximately 50% of all known biologically active SMs from microorganisms), some of which are used in pharmaceutical, agrochemical, and cosmetic products [59]. The majority of these compounds belong to one of four classes obtained through the activity of: (i) nonribosomal peptide synthetase (NRPS), (ii) polyketide synthase (PKS), (iii) terpene cyclase (TC) for terpenoid production, or (iv) a number of enzymes for alkaloid production (Figure 1).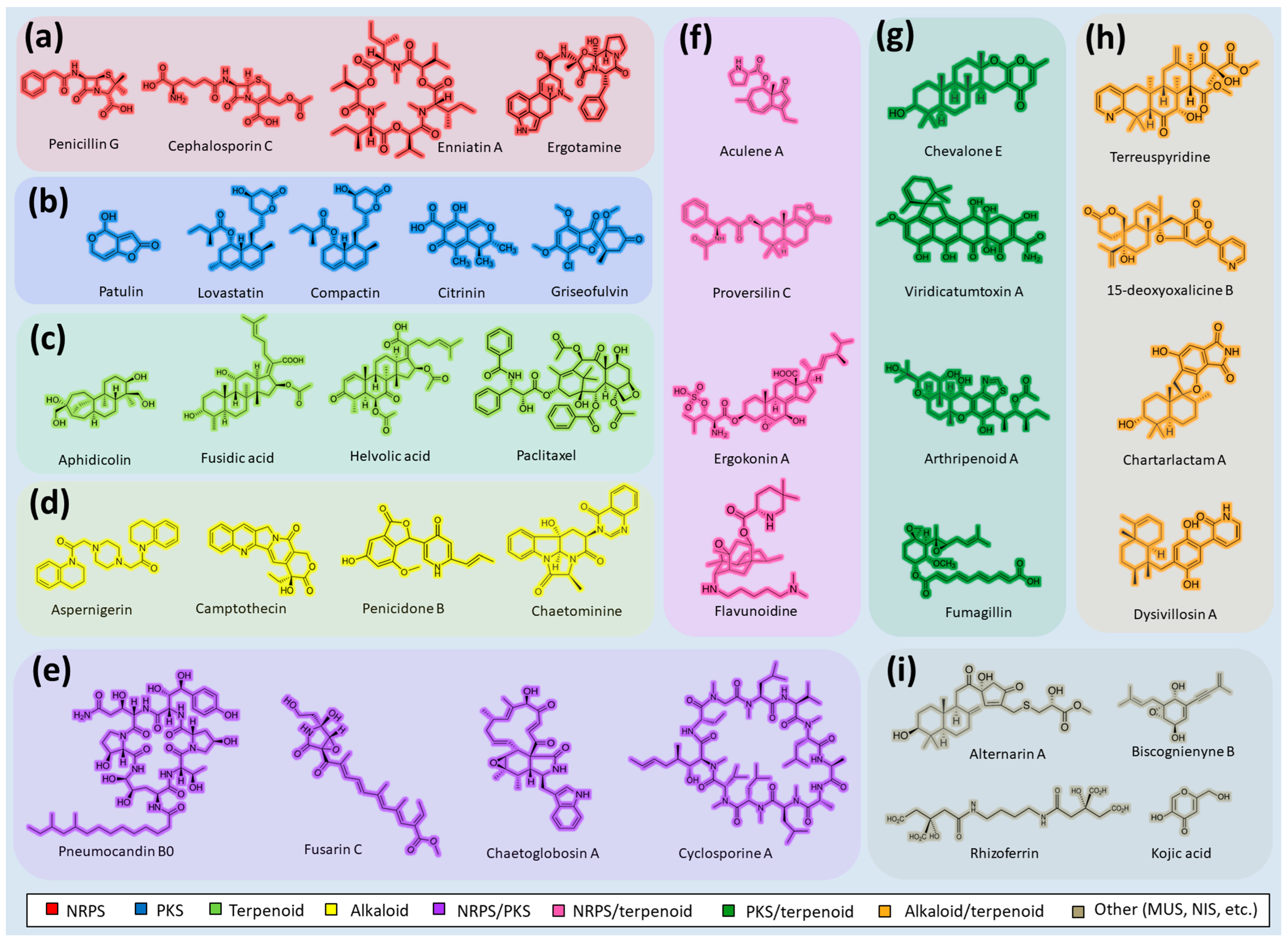
Figure 1. Chemical structures of the main types of secondary metabolites (SMs) produced by filamentous fungi, based on the enzymatic activity of: (a) nonribosomal peptide synthetase (NRPS); (b) polyketide synthase (PKS); (c) terpene cyclase (TPC) for terpenoid production; (d) a number of enzymes for alkaloid production; (e) NRPS and PKS for production of NRPS/PKS hybrid; (f) NRPS and TPC for production of NRPS/terpenoid; (g) PKS and TPC for production of PKS/terpenoid; (h) enzymes for alkaloid production and TPC for production of alkaloid/terpenoid; (i) other enzymes for production meroterpenoid with unique structures (MUS), or NRPS-independent siderophore (NIS), or other types of molecules.
3. Biosynthesis of Fungal Secondary Metabolites in Response to Signals
In most cases, under normal physiological conditions during the trophophase, fungal SMs are not synthesized (Figure 2a) [35]. However, under the influence of certain internal or external signals, cellular mechanisms are triggered, leading to the synthesis of one corresponding (target) SM or another (Figure 2b) [1][15][62].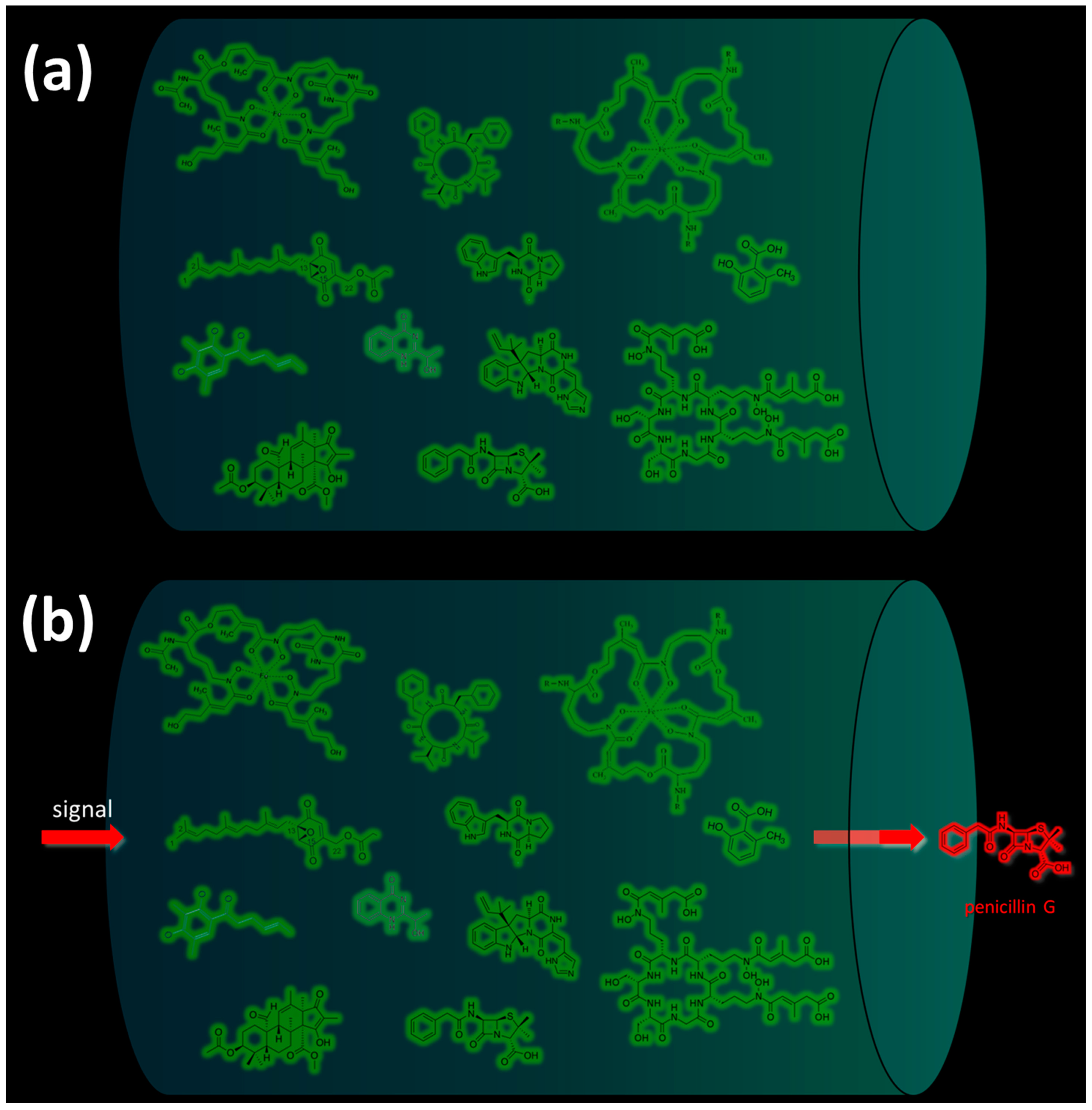
Figure 2. Biosynthesis of secondary metabolites (SMs) in response to signal exposure. The arrival of a specific signal (from the external environment or the internal signal of the cell) leads to the production of corresponding SMs. As an example, changes in the production of SMs in Penicillium chrysogenum are given: (a) Under normal physiological conditions (in the absence of specific environmental signals) and at an early stage of fungal cell development (trophophase stage), most SMs are not produced. (b) In response to a specific signal, the corresponding SM is synthesized. The green color shows known SMs of P. chrysogenum, which, in principle, can be synthesized by the cell (representing its biosynthetic capacity), but are not produced at a particular moment. The red color shows the currently produced SMs in response to the signal; the antibiotic penicillin G, synthesized in response to an external signal, is given as an example.
4. Biosynthetic Gene Clusters (BGCs) for the Production of Fungal Secondary Metabolites
One of the revolutionary discoveries in understanding the molecular basis of the biosynthesis of SMs was the identification of so-called biosynthetic gene clusters (BGCs) [88][89][90][91]. It turned out that in order to create a particular natural product, microorganisms and plants have an appropriate set of genes that are in relative proximity in a particular region of the chromosome (clustered) and are jointly regulated [92]. Thus, the genes responsible for the stages of biosynthesis of a particular SM are either “silent” together or jointly upregulated [26]. The architecture of metabolism itself leads to the maximization of biosynthetic diversity in fungi [93]. For example, a number of BGCs have biased ecological distributions, consistent with niche-specific selection [93]. Several thousand BGCs are currently known in fungi; it is assumed that their numbers range from 100,000 to millions [1][88][94]. There are several main types of BGC organization responsible for the biosynthesis of the corresponding types of SMs in fungi (Figure 1 and Figure 3). In most cases, BGCs contain: (i) one or more genes for backbone, or core, enzymes (synthase or synthetase) responsible for the production of the core structure of SMs, and (ii) a number of genes that encode tailoring enzymes for modifying the core compound to obtain a variety of products [1]. The type of core enzyme (or combination thereof) determines the type of secondary metabolite. The BGC can also assemble genes encoding: (iii) transporters, (iv) proteins that mitigate toxic properties, (v) pathway-specific transcription factors, and (vi) genes with as-yet unknown function (Figure 3) [95].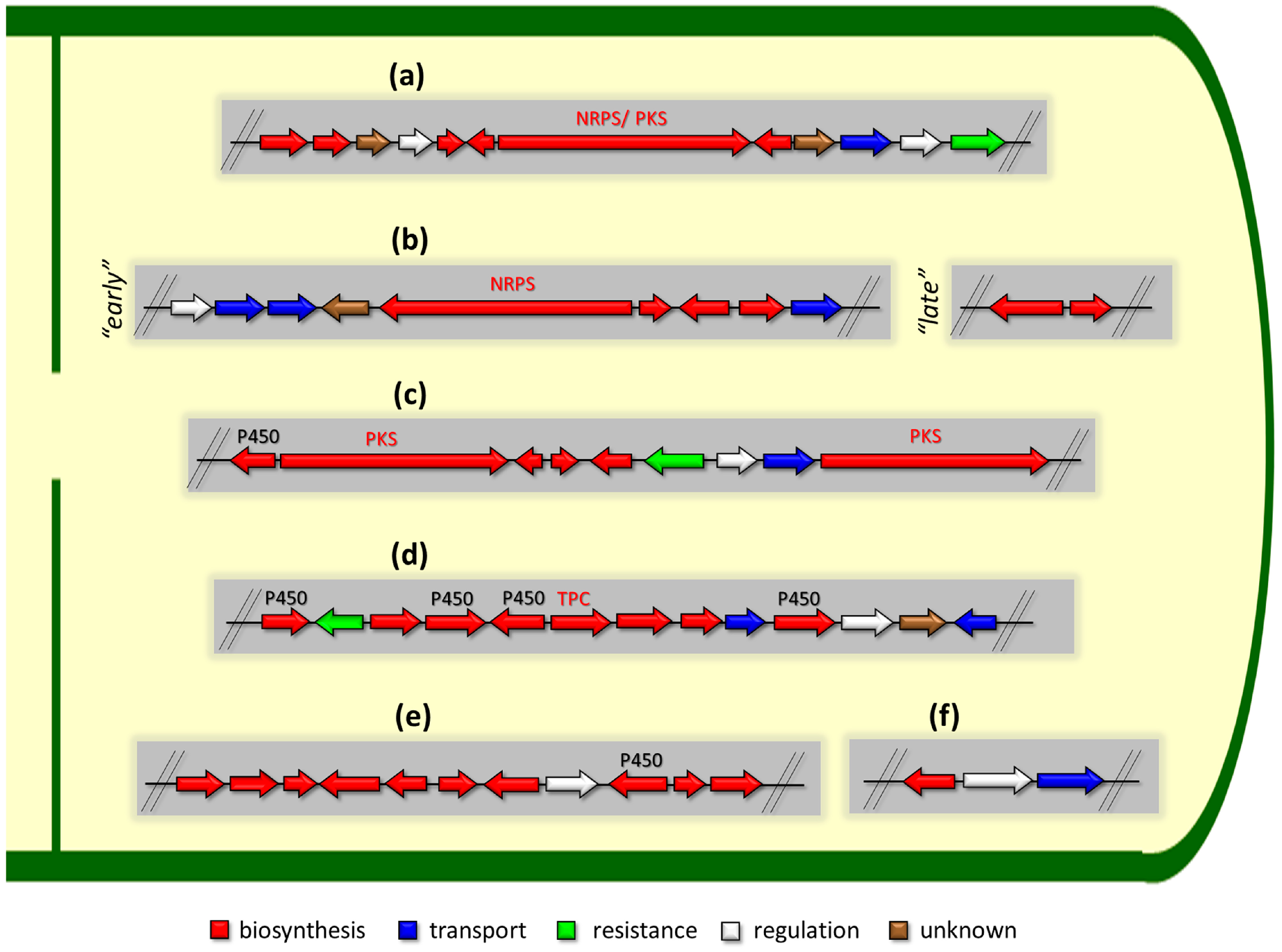
Figure 3. Some examples of the organization of biosynthetic gene clusters (BGCs) for the production of secondary metabolites (SMs) in fungi. (a) BGCs for production of SMs based on so-called “central” gene, which encodes one type of megasynthase or another: (i) NRPS (nonribosomal peptide synthetase), (ii) PKS (polyketide synthase), or (iii) NRPS-PKS hybrid. (b) “Early” and “late” BGCs for production of cephalosporin C in Acremonium chrysogenum. (c) BGCs for production of lovastatin in Aspergillus terreus: P450—cytochrome P450. (d) BGCs for production of terpenoid SM: TPC—terpene cyclase. (e) BGCs for production of meroterpenoid with unique structure. BGC for production of biscognienyne B is given as an example. (f) BGC for production of kojic acid in Aspergillus oryzae. Gene loci for enzymes of the biosynthetic pathways of the SMs are colored in red; gene loci for protein transporters of biosynthetic products are colored in blue; gene locus for protecting the microorganism from the produced secondary metabolite is colored in green; gene locus for the specific regulator of this biosynthetic pathway is colored in white; locus for gene with unknown function is colored in brown. Genes for backbone enzymes (NRPS, PKS, and TPC) responsible for the production of the core structure of SMs are colored in red.
4.1. BGCs with Backbone (or Core) Genes for Megasynthases NRPS or PKS
To create two among the four main types of secondary metabolites, fungi use megasynthases, large modular enzymes such as NRPS, nonribosomal peptide synthetase [96], or PKS (polyketide synthase) [97] (Figure 1a,b and Figure 3a). In these modular enzymes, catalytic domains with a number of functions, required for the polymerization of (i) amino acids, including non-proteinogenic acids (in the case of NRPS), or (ii) acyl groups, from acetyl-CoA to malonyl-CoA (in the case of PKS), are assembled into one huge polypeptide chain [98][99]. As a result, individual megasynthases are responsible for 10–50 or more catalytic activities [15]. In a number of bacteria (~10% of cases), polymerization units do not have a modular organization, and catalytic domains are mainly encoded by individual proteins [100]. It is thought that such non-modular polymerization systems for the production of SMs in bacteria served as a prototype for the development of the modular megasynthases NRPS and PKS [100]. Each module of NRPS is a functional building block responsible for incorporating and modifying a single amino acid unit, which can be either canonical proteinogenic (i.e., used in ribosomal synthesis) or non-canonical non-proteinogenic (i.e., never used in ribosomal synthesis) [101][102]. A typical NRPS module consists of: (i) the adenylation (A) domain, for amino acid recognition and activation; (ii) the peptidyl carrier protein (PCP) domain, for transferring an activated amino acid from the A-domain to its cofactor, 4′-phosphopantetheine; and (iii) the condensation (C) domain, to catalyze peptide bond formation [103]. Along with this, the module may contain a set of optional domains with catalytic functions of methyltransferase (MT), β-ketoacyl reductase (KR), epimerase (E), etc. [104]. Specificity of the recognition of one amino acid or another is achieved due to the substrate-binding center of the adenylation domain of the corresponding module [105][106]. In this regard, the term “nonribosomal” code was introduced, referring to the correspondence of 10 amino acid residues in the substrate binding site of the adenylation domain of a particular NRPS module with a specific proteinogenic or non-proteinogenic amino acid [107][108]. More than 500 non-proteinogenic amino acids have now been found in fungi, many of which are used for non-ribosomal peptide synthesis [109][110]. In addition, for the biosynthesis of a number of non-proteinogenic amino acids themselves, an additional BGC is required [111]. Adding such a significant number of “building block” types to the canonical 20 proteinogenic amino acids (the number of which is strictly limited by genetic coding and the rigidly fixed roles of tRNA and aminoacyl-tRNA synthetizes) makes it possible to drastically expand the range of created low-molecular-weight structures [109]. Fundamentally new structures emerging as a result of the use of new building materials on the NRPS platform provide an advantage to the organisms that produce them, and can also be applied to obtain medically significant natural products [112][113][114]. PKS can have, as in the case of NRPS, a complex multi-module structure (type I noniterative PKS) where a single module from a huge enzyme with multiple modules is used to attach the next building block [115][116]. Such enzymes function as a modular linear conveyor line, in which each active site is used only once [117]. However, in fungi, the most common PKS is the iterative type (type I iterative PKS and type II PKS), which, instead of one large megaenzyme, consists of only one module that reuses necessary catalytic domains in a cyclic fashion [118]. After attaching a building block, the polymerization product is transferred to the beginning of the module to attach the next building block, and so on [119]. Such enzymes function as an iterative assembly line in which each active site of the core domains is used as many times as needed to attach the building blocks [120]. Typically, a single PKS module contains three core (minimal) domains: (i) the acyl transferase (AT) domain selects the building blocks to add to the product and transfers them to (ii) the acyl transfer protein (ACP) domain, which loads them for the polymerization product, and (iii) the ketoacyl synthase (KS) domain, which is required for the decarboxylation condensation of the extendable unit (usually malonyl-CoA or methylmalonyl-CoA) with the acyl thioether [121]. There is also iterative AT-less and ACP-less type III PKS in fungi, which is a homodimer with a molecular weight of about 40 kDa and combines all the activities from the essential type I and II PKS domains [122]. Along with minimal domains, the module may contain a set of optional (or tailoring) domains with catalytic functions of thioesterase (TE), methyltransferase (MT), dehydratase (DH), enoyl reductase (ER), β-ketoacyl reductase (KR), etc. [121]. Depending on the presence and number of reducing domains in PKS, they are subdivided into: (i) NR-PKS—non-reducing PKS, the products of which are true polyketides; (ii) PR-PKS—partially contracting PKS; and (iii) FR-PKS—fully reducing PKS, the products of which are fatty acid derivatives. As a result of this diversity of intramodular organization, PKS, along with NRPS, produce an enormously diverse array of natural products in fungi [123]. There are also known cases when more than one corresponding megasynthase is used for the production of NRPS-driven (Figure 1a) or PKS-driven (Figure 1b) secondary metabolites by fungi. For example, two PKSs are used during lovastatin biosynthesis, one of which, LovB nonaketide synthase (EC:2.3.1.161), uses nine building blocks based on acetyl-CoA or manoyl-CoA, and the other, LovF diketide synthase (2-methylbutanoate polyketide synthase; EC: 2.3.1.244), uses two such building blocks [124]. Accordingly, the lovastatin BGC encodes two PKS genes (Figure 3c). There are also numerous examples of BGCs in fungi encoding both NRPS and PKS.4.2. BGCs with Backbone (or Core) Gene for Terpene Cyclase
Terpene cyclase (TPC) is used as the core enzyme for the biosynthesis of the third among the four major types of fungal secondary metabolites, terpenoids (Figure 1c and Figure 3d) [125]. In most cases, TPC clusters in the same BGC as its downstream modification enzymes (Figure 3d) [126]. TPCs form the hydrocarbon backbones of terpenoids, which are then modified by tailoring enzymes to produce final natural products [127]. Depending on the initial generation of the carbocation, class I TPK and class II TPK are distinguished [128]. TPC is a catalytic complex that produces cyclic terpenoids from their linear precursors [129]. Terpenoid cyclization reactions are one of the most complex reactions found in nature [130]. Due to the functional diversity of terpene cyclases, various types of cyclic terpenoids are formed from linear precursors, which, in turn, undergo various modifications. Currently, over 80,000 terpenoids are known, which represent about a third of the described natural products [131]. In most cases, the gene for TPC clusters in the same BGC as the genes for its downstream modification enzymes [126]. However, there are a number of examples, such as lanosterol-derived triterpenes/steroids, where the TPC gene is outside the gene cluster for its downstream modification enzymes [132].4.3. Hybrid BGCs with Genes for Different Backbone Enzymes
In addition to biosynthetic clusters encoding only one type of core enzyme, which leads, respectively, to the production of secondary metabolites of the NRPS type, PKS type, or TPC type (Figure 1b–d), there are mixed-type BGCs that contain genes for different types of core enzymes [88]. There are also BGCs with hybrid core genes, for example, for the production of NRPS/PKS hybrids, part of the gene may encode NRPS modules and the other part PKS modules [101]. In such cases, specific interpolypeptide linkers exist at both the C- and N-termini of the NRPS and PKS proteins, which play a critical role in facilitating the transfer of the growing peptide or polyketide intermediate between NRPS and PKS modules in hybrid NRPS-PKS systems [88]. Among the four basic types of SMs in fungi (NRPS, PKS, terpenes, and alkaloids), there are numerous chimeric variants. As a result, the production of such mixed (or hybrid) fungal BGCs results in chimeric secondary metabolites such as NRPS/PKS, NRPS/terpenoid, PKS/terpenoid, or alkaloid/terpenoid hybrids (Figure 1e–h) [60][88][133]. Some (but not all) alkaloids also use core enzymes for their construction [134][135]; for example, ergot alkaloids use NRPS [136][137]. In rare cases, secondary metabolites in fungi result from crosstalk between two separate BGCs [138]. Such an interaction not only increases the structural diversity but also significantly expands the activity spectrum of the produced cross-cluster compounds [138]. NRPS-PKS hybrids (Figure 1e) are among the most common in nature [139]. Such compounds benefit from the combinatorics of products resulting from NRPS and PKS synthesis [101]. It has been shown that more than a third of the clusters encoding megasynthases carry NRPS-PKS hybrids [100].4.4. BGCs without Genes for Canonical Backbone Enzymes (“Wild BGCs”)
In addition to the main types of SMs, in the production of which relatively easily identifiable genes of core and tailoring enzymes are involved (Figure 1a–h), fungi also produce highly active low-molecular-weight compounds that do not have characteristic elements for their “barcoding” (Figure 1i) [140]. BGCs for the production of such SMs do not contain genes encoding canonical “backbone” synthases/synthetases (e.g., NRPS, PKS, TPC); for example, clusters for the production of clavine alkaloids [141], isocyanides [142], NRPS-independent siderophores (NIS) [143], and other [144]. BGCs for the production of clavine alkaloids do not contain NRPS [141], unlike ergot alkaloids, with four genes encoding NRPS [145]. Isocyanides (also called isonitriles) have notable bioactivities that mediate pathogenesis, microbial competition, and metal homeostasis through metal-associated chemistry [142]. For isocyanide production, fungi use non-canonical BGCs (containing the non-canonical core enzyme isocyanide synthase, ICS), which are not detected by standard genome-mining algorithms [146]. However, a targeted bioinformatics study of 3300 fungal genomes allowed 3800 ICS BGCs to be characterized [140]. Hydroxamic siderophores also use NRPS, but recently, an NRPS-independent siderophore (NIS) synthetase pathway has been established for the production of NRPS-independent siderophores [147]. Five functional types of NIS enzymes are classified; all such clusters also lack the core canonical gene [148]. The BGC for kojic acid production does not contain genes encoding both core enzymes and characteristic tailoring enzymes (Figure 3f) [144]. The lack of conserved signature sequences makes such BGCs almost impossible to detect as a result of genomic mining using current bioinformatic approaches [149]. The only way to detect such clusters is through an experimental approach. For example, the BGC of kojic acid in Aspergillus oryzae was identified as a result of a reverse genetic method combined with a DNA microarray technique [144]. Currently, most of ourthe knowledge about BGCs is formed in silico [150]. As a result of the application of bioinformatics technologies, tens of thousands of BGCs have been found in fungal genomes, for most of which the products are still unknown [88]. Along with this, for all secondary metabolites from bacteria, fungi, and plants, fewer than two thousand corresponding BGCs have been experimentally characterized [151][152]. As a result, ourthe knowledge of “wild” clusters (without characteristic core and tailoring enzymes) is much narrower than that of BGCs containing these elements. There are also “canonical” BGCs without genes for core enzymes. This is due to the fact that genes encoding canonical core enzymes for such clusters are localized outside the cluster, in the other part of the genome. For example, the “late” beta-lactam BGC contains only genes for tailoring enzymes (CefEF and CefG), while the core enzyme for this biosynthetic pathway clusters in the “early” beta-lactam BGC, which is located on a different chromosome (Figure 3b) [153][154].4.5. Tailoring Enzymes (Enzymes for Modifying the Core Structure)
The final products of biosynthetic secondary metabolism pathways are often significantly modified as a result of enzymatic activities such as heterocyclization, epimerization, oxidative hydroxylation, methylation, oxidative crosslinking, the addition of sugars, translocation, and other modifications [155]. Some tailoring enzymes assemble as optional domains within megasynthase modules; other tailoring enzymes act in trans during megasynthase work, recognizing the modules required by protein–protein interactions [155]. For example, the trans-acting polyketide enoyl reductase LovC (lovastatin enoyl reductase; EC: 2.3.1.161) specifically reduces three out of eight polyketide intermediates (triketides, tetraketides, and hexaketides) during nonaketide synthase LovB activity in lovastatin biosynthesis [124]. As a result of such cis- and trans-activities, the core polymerization product may contain, after the release, a significant number of modifications. The release of the core scaffold process itself is quite complex; it can proceed using various mechanisms [156], the implementation of which may also require special enzymes encoded in the corresponding BGC. For example, in the biosynthesis of lovastatin, thioester hydrolases LovG (dihydromonacolin L-[lovastatin nonaketide synthase] thioesterase; EC: 3.1.2.31) is required to release from nonaketide synthase LovB its final product, dihydromonacolin L [157]. After the backbone, or core, enzymes create a core scaffold (with cis- and possibly trans-modifications), a third group of tailoring enzymes transform its structure, resulting in a variety of end products. Thus, in addition to the genes for core enzymes, BGCs contain genes for various biosynthetic enzymes, trans-acting with core enzymes, helping to release or modify the released core products (Figure 3). For example, in A. chrysogenum, after NRPS, which is called PcbAB or ACV (δ-[L-α-Aminoadipoyl]-L-Cysteinyl-D-Valine) synthetase (EC: 6.3.2.26), polymerizes the LLD-ACV tripeptide δ-(L-α-aminoadipoyl)-L-cysteinyl-D-valine, a series of enzymatic reactions occur, catalyzed by enzymes from beta-lactam BGCs, resulting in the production of cephalosporin C (CPC). First, PcbC (isopenicillin N-synthase (EC: 1.21.3.1)), as a result of a dioxygenase reaction, cyclizes this tripeptide to isopenicillin N (IPN); then, cefD1 (isopenicillin N-CoA synthetase (EC: 5.1.1.17)), and cefD2 (isopenicillin N-CoA epimerase (EC: 5.1.1.17)) catalyze reactions leading to the epimerization of IPN to penicillin N (penN); finally, enzymes of the “late” beta-lactam BGC, CefEF (deacetoxycephalosporin C synthetase (penicillin N expandase, EC: 1.14.20.1)/deacetoxycephalosporin C hydroxylase (EC: 1.14.11.26)), and CefG (deacetylcephalosporin-C acetyltransferase (EC: 2.3. 1.175)), carry out reactions leading to the formation of CPC [158][159][160]. A distinctive feature of BGC in terpenoid biosynthesis is the presence among the genes for tailoring enzymes of a significant number of genes for cytochrome P450 mono-oxygenases (CYP450), NAD(P)+, and flavin-dependent oxidoreductases that generate the final bioactive structures (Figure 3d) [55]. Individual members of the CYP450 superfamily catalyze various stereospecific modifications at various positions in the core structures of terpenoids, as a result of which their biological activity can significantly increase [161][162]. The most important modification catalyzed by CYP450 is oxidative hydroxylation, which makes the compound more hydrophilic [163]. Clustered NAD(P)+ and flavin-dependent oxidoreductases are required for CYP450 to function as partners in the electron transfer chain [164]. In addition to terpenoids, CYP450s are also used to modify other types of fungal secondary metabolites based on NRPS, PKS, and NRPS-PKS activities and meroterpenoids [165]. For example, LovA (CYP68R1, dihydromonacolin L/monacolin L hydroxylase; EC: 1.14.14.124, EC: 1.14.14.125) from the lovastatin biosynthetic pathway sequentially introduces two hydroxyl groups into the backbone (dihydromonacolin L), which leads to: (i) the introduction of the 4a,5-double bond and obtaining monacolin L, which, in turn, (ii) is hydroxylated at C-8 to form monacolin J [166]. The hydroxyl inserted at the C-8 position is then used to incorporate the independently synthesized diketide via a transferase reaction involving LovD (monacolin-J-acid methylbutanoate transferase; EC: 2.3.1.238) to form the final product, lovastatin [167]. However, CYP450s localized separately (without association with any core enzyme of VM biosynthesis) are not always good indicators for the search for biosynthetic clusters of secondary metabolism, since they are used not only to build secondary metabolism, but also for the biosynthesis of structural components and in signaling networks, and are instrumental in xenobiotic detoxification [161][168][169][170]. There are currently about 400 CYP families (namely, CYP51-CYP69, CYP501-CYP699, and CYP5001-CYP6999) [171], which exceeds the diversity in the number of families of representatives of this protein superfamily in bacteria (333 CYP families), plants (127 CYP families), vertebrates (19 CYP families), and insects (67 CYP families) [161]. Due to this variety in the most important enzymatic components of fungi, as well as the lack of data on structural and functional relationships for the vast majority of CYP450, the presence of their genes is only a signal for a possible search for BGCs.4.6. Transporter Genes of BGCs
It also turns out that, together with the genes for the biosynthesis of a secondary metabolite, the genes necessary for the transport of the final product or its intermediates can be clustered [26][158][172][173]. Such transport can occur both for the removal of the end product from the cell, and for the transport of metabolic intermediates between different compartments of the cell, where the stages of biosynthesis take place [158][174][175][176]. For example, in A. chrysogenum, the first steps in the biosynthesis of cephalosporin C (CPC), leading to the biosynthesis of IPN, occur in the cytoplasm; then, in the peroxisome, epimerization of IPN to penicillin N (penN) occurs; the final conversion of penN into the target SM, CPC, occurs again in the cytoplasm [177]. For this purpose, in the “early” BGC of beta-lactams, there are special genes for transporter proteins that carry out active transport of the corresponding intermediates: first, as a result of the activity of the CefP transporter, IPN enters peroxisome from the cytoplasm [178]; then reactions occur in the peroxisome, leading to the epimerization of IPN to PenG [159], which then, as a result of the activity of the CefM transporter [179], moves from the peroxisome to the cytoplasm, where it undergoes further transformations.4.7. Gene for Resistance of BGCs
Another important class of genes found in BGCs are resistance genes against the directly synthesized compound (Figure 3). The physiological basis of this strategy is that many high-yielding natural products, such as antibiotics or statins, can harm the host organism by acting on microorganisms with similar biochemistry [1]. This is why it is necessary to “defend” against a number of compounds created by the microorganism itself [174][180]. Currently, three main defense strategies for BGC resistance genes have been classified. They are associated with: (i) placement in the BGC of an additional copy of the gene encoding the target protein, which is inhibited by the produced metabolite; (ii) the active transport of a “hazardous” substance from the cell; and (iii) the coding of an enzyme that detoxifies the final highly active antimicrobial product [70]. For example, the “early” beta-lactam BGC also contains the gene for the CefT transporter, which serves in the active transport of CPC and its intermediates, such as IPN, PenN, deacetoxycephalosporin C (DAOC), and deacetylcephalosporin C (DAC), out of the cell [158][181]. In the BGC for the production of lovastatin (LOV), a compound that affects the ergosterol biosynthesis of competing fungi (and potentially affects endogenous ergosterol biosynthesis), lovR is clustered, representing an additional copy of the gene encoding 3-hydroxy-3-methyl glutaryl coenzyme A reductase (EC: 1.1.1.34), which is inactivated by LOV as a result of irreversible binding.4.8. Pathway-Specific and Cross-Cluster Regulators of BGCs
Finally, in addition to genes for biosynthesis, transport, and resistance, there is a fourth class of genes, often, but not always, found in BGCs, that are responsible for pathway-specific regulation of the BGC itself and/or of other BGCs, in the case of cross-regulation [94]. Such genes encode transcription factors that are able to modulate the effect of signals perceived and reproduced by global regulators and occur in more than half of the currently known BGCs [182]. These factors can act as positive regulators during the signal amplification stage [94]. There are also negative pathway-specific regulators leading to downregulation of the BGC; they are more common if genes for two regulators are clustered in the same BGC and one regulator is positive while the other is negative [94]. However, there are regulators that can be positive for some BGC genes and negative for others. For example, in A. chrysogenum, the early BGC beta-lactam cluster contains a gene for the CefR regulator, which is both a negative regulator for the cefT transporter gene from the early BGC and a positive regulator for the cefEF biosynthetic gene from the late BGC [183]. Thus, CefR from the early BGC beta-lactam cluster is a pathway-specific regulator for cefT, and a cross-cluster regulator for cefEF. Such a differential effect of CefR on the expression of beta-lactam BGCs in A. chrysogenum allows, on the one hand, the biosynthesis of CPC to be intensified (as a result of upregulation of one of the key biosynthetic genes), and on the other hand, for a reduction in the “leakage” of intermediates from the cell (such as IPN, PenN, DAOC, and DAC) and their redirection toward producing the target metabolite, CPC.References
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180.
- Bayer, E.A.; Fierro, F.; Vaca, I.; Castillo, N.I.; Ovidio García-Rico, R.; Chávez, R. Penicillium chrysogenum, a Vintage Model with a Cutting-Edge Profile in Biotechnology. Microorganisms 2022, 10, 573.
- Boruta, T. Uncovering the repertoire of fungal secondary metabolites: From Fleming’s laboratory to the International Space Station. Bioengineered 2017, 9, 12–16.
- Aly, A.H.; Debbab, A.; Proksch, P. Fifty years of drug discovery from fungi. Fungal Divers. 2011, 50, 3–19.
- Avalos, J.; Limón, M.C. Fungal Secondary Metabolism. Encyclopedia 2021, 2, 1–13.
- Brown, R.; Priest, E.; Naglik, J.R.; Richardson, J.P. Fungal Toxins and Host Immune Responses. Front. Microbiol. 2021, 12, 643639.
- Maresca, M.; Muchagato Mauricio, E.; Brito, L.; Conrado, R.; Colombo Gomes, T.; Sales Calaço Roque, G.; Olívia De Souza, A. Overview of Bioactive Fungal Secondary Metabolites: Cytotoxic and Antimicrobial Compounds. Antibiotics 2022, 11, 1604.
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 2010, 8, 423–435.
- Mazzei, T.; Mini, E.; Novelli, A.; Periti, P. Chemistry and mode of action of macrolides. J. Antimicrob. Chemother. 1993, 31, 1–9.
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260.
- Bush, K.; Bradford, P.A. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247.
- Fàbrega, A.; Madurga, S.; Giralt, E.; Vila, J. Mechanism of action of and resistance to quinolones. Microb. Biotechnol. 2009, 2, 40–61.
- Blondeau, J.M. Fluoroquinolones: Mechanism of action, classification, and development of resistance. Surv. Ophthalmol. 2004, 49, S73–S78.
- Kotra, L.P.; Haddad, J.; Mobashery, S. Aminoglycosides: Perspectives on Mechanisms of Action and Resistance and Strategies to Counter Resistance. Antimicrob. Agents Chemother. 2000, 44, 3249–3256.
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32.
- Rokas, A.; Mead, M.E.; Steenwyk, J.L.; Raja, H.A.; Oberlies, N.H. Biosynthetic gene clusters and the evolution of fungal chemodiversity. Nat. Prod. Rep. 2020, 37, 868–878.
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; De Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631.
- Scherlach, K.; Hertweck, C. Mining and unearthing hidden biosynthetic potential. Nat. Commun. 2021, 12, 3864.
- Gilchrist, C.L.M.; Li, H.; Chooi, Y.H. Panning for gold in mould: Can we increase the odds for fungal genome mining? Org. Biomol. Chem. 2018, 16, 1620–1626.
- Gressler, M.; Zaehle, C.; Scherlach, K.; Hertweck, C.; Brock, M. Multifactorial induction of an orphan PKS-NRPS gene cluster in Aspergillus terreus. Chem. Biol. 2011, 18, 198–209.
- Brakhage, A.A.; Schroeckh, V. Fungal secondary metabolites—Strategies to activate silent gene clusters. Fungal Genet. Biol. 2011, 48, 15–22.
- Hannigan, G.D.; Prihoda, D.; Palicka, A.; Soukup, J.; Klempir, O.; Rampula, L.; Durcak, J.; Wurst, M.; Kotowski, J.; Chang, D.; et al. A deep learning genome-mining strategy for biosynthetic gene cluster prediction. Nucleic Acids Res. 2019, 47, e110.
- Yee, D.A.; Niwa, K.; Perlatti, B.; Chen, M.; Li, Y.; Tang, Y. Genome mining for unknown–unknown natural products. Nat. Chem. Biol. 2023, 19, 633–640.
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes—A review. Nat. Prod. Rep. 2016, 33, 988–1005.
- Kjærbølling, I.; Mortensen, U.H.; Vesth, T.; Andersen, M.R. Strategies to establish the link between biosynthetic gene clusters and secondary metabolites. Fungal Genet. Biol. 2019, 130, 107–121.
- Mózsik, L.; Iacovelli, R.; Bovenberg, R.A.L.; Driessen, A.J.M. Transcriptional Activation of Biosynthetic Gene Clusters in Filamentous Fungi. Front. Bioeng. Biotechnol. 2022, 10, 901037.
- Wang, W.; Drott, M.; Greco, C.; Luciano-Rosario, D.; Wang, P.; Keller, N.P. Transcription Factor Repurposing Offers Insights into Evolution of Biosynthetic Gene Cluster Regulation. MBio 2021, 12, 10–1128.
- Baral, B.; Akhgari, A.; Metsä-Ketelä, M. Activation of microbial secondary metabolic pathways: Avenues and challenges. Synth. Syst. Biotechnol. 2018, 3, 163–178.
- Brakhage, A.A.; Spröte, P.; Al-Abdallah, Q.; Gehrke, A.; Plattner, H.; Tüncher, A. Regulation of penicillin biosynthesis in filamentous fungi. Adv. Biochem. Eng. Biotechnol. 2004, 88, 45–90.
- Yu, J.H.; Keller, N. Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 2005, 43, 437–458.
- Kosalková, K.; García-Estrada, C.; Ullán, R.V.; Godio, R.P.; Feltrer, R.; Teijeira, F.; Mauriz, E.; Martín, J.F. The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine C synthesis in Penicillium chrysogenum. Biochimie 2009, 91, 214–225.
- Yin, W.; Keller, N.P. Transcriptional regulatory elements in fungal secondary metabolism. J. Microbiol. 2011, 49, 329–339.
- Bind, S.; Bind, S.; Sharma, A.K.; Chaturvedi, P. Epigenetic Modification: A Key Tool for Secondary Metabolite Production in Microorganisms. Front. Microbiol. 2022, 13, 784109.
- Calvo, A.M.; Wilson, R.A.; Bok, J.W.; Keller, N.P. Relationship between Secondary Metabolism and Fungal Development. Microbiol. Mol. Biol. Rev. 2002, 66, 447–459.
- Demain, A.L. Regulation of secondary metabolism in fungi. Pure Appl. Chem. 1986, 58, 219–226.
- Macheleidt, J.; Mattern, D.J.; Fischer, J.; Netzker, T.; Weber, J.; Schroeckh, V.; Valiante, V.; Brakhage, A.A. Regulation and Role of Fungal Secondary Metabolites. Annu. Rev. Genet. 2016, 50, 371–392.
- Haas, H. How to trigger a fungal weapon. eLife 2015, 4, e10504.
- Gressler, M.; Meyer, F.; Heine, D.; Hortschansky, P.; Hertweck, C.; Brock, M. Phytotoxin production in Aspergillus terreus is regulated by independent environmental signals. eLife 2015, 4, e07861.
- García-Estrada, C.; Martín, J.F.; Cueto, L.; Barreiro, C. Omics Approaches Applied to Penicillium chrysogenum and Penicillin Production: Revealing the Secrets of Improved Productivity. Genes 2020, 11, 712.
- Shankar, A.; Sharma, K.K. Fungal secondary metabolites in food and pharmaceuticals in the era of multi-omics. Appl. Microbiol. Biotechnol. 2022, 106, 3465–3488.
- Barreiro, C.; García-Estrada, C. Proteomics and Penicillium chrysogenum: Unveiling the secrets behind penicillin production. J. Proteom. 2019, 198, 119–131.
- Zhgun, A.A.; Eldarov, M.A. Polyamines Upregulate Cephalosporin C Production and Expression of β-Lactam Biosynthetic Genes in High-Yielding Acremonium chrysogenum Strain. Molecules 2021, 26, 6636.
- Domratcheva, A.G.; Zhgun, A.A.; Novak, N.V.; Dzhavakhiya, V.V. The Influence of Chemical Mutagenesis on the Properties of the Cyclosporine a High-Producer Strain Tolypocladium inflatum VKM F-3630D. Appl. Biochem. Microbiol. 2018, 54, 53–57.
- Zhgun, A.A.; Nuraeva, G.K.; Dumina, M.V.; Voinova, T.M.; Dzhavakhiya, V.V.; Eldarov, M.A. 1,3-Diaminopropane and Spermidine Upregulate Lovastatin Production and Expression of Lovastatin Biosynthetic Genes in Aspergillus terreus via LaeA Regulation. Appl. Biochem. Microbiol. 2019, 55, 243–254.
- Bladt, T.T.; Frisvad, J.C.; Knudsen, P.B.; Larsen, T.O. Anticancer and Antifungal Compounds from Aspergillus, Penicillium and Other Filamentous Fungi. Molecules 2013, 18, 11338–11376.
- Weber, S.S.; Bovenberg, R.A.L.; Driessen, A.J.M. Biosynthetic concepts for the production of β-lactam antibiotics in Penicillium chrysogenum. Biotechnol. J. 2012, 7, 225–236.
- Bunch, A.W.; Harris, R.E. The manipulation of micro-organisms for the production of secondary metabolites. Biotechnol. Genet. Eng. Rev. 1986, 4, 117–144.
- Wu, M.; Crismaru, C.G.; Salo, O.; Bovenberg, R.A.L.; Driessena, A.J.M. Impact of Classical Strain Improvement of Penicillium rubens on Amino Acid Metabolism during β-Lactam Production. Appl. Environ. Microbiol. 2020, 86, e01561-19.
- Salo, O.V.; Ries, M.; Medema, M.H.; Lankhorst, P.P.; Vreeken, R.J.; Bovenberg, R.A.L.; Driessen, A.J.M. Genomic mutational analysis of the impact of the classical strain improvement program on β-lactam producing Penicillium chrysogenum. BMC Genom. 2015, 16, 937.
- Nielsen, J.C.; Nielsen, J. Development of fungal cell factories for the production of secondary metabolites: Linking genomics and metabolism. Synth. Syst. Biotechnol. 2017, 2, 5–12.
- Guzmán-Chávez, F.; Zwahlen, R.D.; Bovenberg, R.A.L.; Driessen, A.J.M. Engineering of the filamentous fungus Penicillium chrysogenum as cell factory for natural products. Front. Microbiol. 2018, 9, 2768.
- Tedersoo, L.; May, T.W.; Smith, M.E. Ectomycorrhizal lifestyle in fungi: Global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 2010, 20, 217–263.
- Galagan, J.E.; Henn, M.R.; Ma, L.J.; Cuomo, C.A.; Birren, B. Genomics of the fungal kingdom: Insights into eukaryotic biology. Genome Res. 2005, 15, 1620–1631.
- Blackwell, M. The fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 2011, 98, 426–438.
- Quin, M.B.; Flynn, C.M.; Schmidt-Dannert, C. Traversing the fungal terpenome. Nat. Prod. Rep. 2014, 31, 1449–1473.
- Goga, M.; Elečko, J.; Marcinčinová, M.; Ručová, D.; Bačkorová, M.; Bačkor, M. Lichen Metabolites: An Overview of Some Secondary Metabolites and Their Biological Potential. In Co-Evolution of Secondary Metabolites; Springer: Cham, Switzerland, 2020; pp. 175–209.
- Molnár, K.; Farkas, E. Current results on biological activities of lichen secondary metabolites: A review. Z. Naturforsch. C. 2010, 65, 157–173.
- Singh, G. Linking Lichen Metabolites to Genes: Emerging Concepts and Lessons from Molecular Biology and Metagenomics. J. Fungi 2023, 9, 160.
- Bills, G.F.; Gloer, J.B. Biologically Active Secondary Metabolites from the Fungi. Microbiol. Spectr. 2016, 4, 4–6.
- Han, J.; Jiang, L.; Zhang, L.; Quinn, R.J.; Liu, X.; Feng, Y. Peculiarities of meroterpenoids and their bioproduction. Appl. Microbiol. Biotechnol. 2021, 105, 3987–4003.
- Carroll, C.S.; Moore, M.M. Ironing out siderophore biosynthesis: A review of non-ribosomal peptide synthetase (NRPS)-independent siderophore synthetases. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 356–381.
- Kumar, A.; Kumar, A. Synthesis and Regulation of Fungal Secondary Metabolites. Microb. Technol. Welf. Soc. 2019, 17, 25–52.
- Netzker, T.; Fischer, J.; Weber, J.; Mattern, D.J.; König, C.C.; Valiante, V.; Schroeckh, V.; Brakhage, A.A. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front. Microbiol. 2015, 6, 299.
- Kai, K. Bioorganic chemistry of signaling molecules in microbial communication. J. Pestic. Sci. 2019, 44, 200–207.
- Abdul Hamid, N.W.; Nadarajah, K. Microbe Related Chemical Signalling and Its Application in Agriculture. Int. J. Mol. Sci. 2022, 23, 8998.
- Oppong-Danquah, E.; Budnicka, P.; Blümel, M.; Tasdemir, D. Design of Fungal Co-Cultivation Based on Comparative Metabolomics and Bioactivity for Discovery of Marine Fungal Agrochemicals. Mar. Drugs 2020, 18, 73.
- Gerke, J.; Köhler, A.M.; Wennrich, J.-P.; Große, V.; Shao, L.; Heinrich, A.K.; Bode, H.B.; Chen, W.; Surup, F.; Braus, G.H. Biosynthesis of Antibacterial Iron-Chelating Tropolones in Aspergillus nidulans as Response to Glycopeptide-Producing Streptomycetes. Front. Fungal Biol. 2022, 2, 68.
- Chevrette, M.G.; Thomas, C.S.; Hurley, A.; Rosario-Melendez, N.; Sankaran, K.; Tu, Y.; Hall, A.; Magesh, S.; Handelsman, J. Microbiome composition modulates secondary metabolism in a multispecies bacterial community. Proc. Natl. Acad. Sci. USA 2022, 119, e2212930119.
- Demain, A.L.; Fang, A. The natural functions of secondary metabolites. Adv. Biochem. Eng. Biotechnol. 2000, 69, 1–39.
- Keller, N.P. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol. 2015, 11, 671–677.
- Scharf, D.H.; Heinekamp, T.; Brakhage, A.A. Human and Plant Fungal Pathogens: The Role of Secondary Metabolites. PLoS Pathog. 2014, 10, e1003859.
- Thines, E.; Aguirre, J.; Foster, A.J.; Deising, H.B. Genetics of Phytopathology: Secondary Metabolites as Virulence Determinants of Fungal Plant Pathogens. In Progress in Botany; Springer: Berlin/Heidelberg, Germany, 2006; pp. 134–161.
- Mapuranga, J.; Chang, J.; Zhang, L.; Zhang, N.; Yang, W.; Kononowicz, A.K.; Macioszek, V.K.; Mapuranga, J.; Chang, J.; Zhang, L.; et al. Fungal Secondary Metabolites and Small RNAs Enhance Pathogenicity during Plant-Fungal Pathogen Interactions. J. Fungi 2022, 9, 4.
- Pusztahelyi, T.; Holb, I.J.; Pócsi, I. Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 2015, 6, 573.
- Rohlfs, M.; Churchill, A.C.L. Fungal secondary metabolites as modulators of interactions with insects and other arthropods. Fungal Genet. Biol. 2011, 48, 23–34.
- Vargas, W.A.; Mukherjee, P.K.; Laughlin, D.; Wiest, A.; Moran-Diez, M.E.; Kenerley, C.M. Role of gliotoxin in the symbiotic and pathogenic interactions of Trichoderma virens. Microbiology 2014, 160, 2319–2330.
- Mehmood, A.; Liu, G.; Wang, X.; Meng, G.; Wang, C.; Liu, Y. Fungal Quorum-Sensing Molecules and Inhibitors with Potential Antifungal Activity: A Review. Molecules 2019, 24, 1950.
- Dufour, N.; Rao, R.P. Secondary metabolites and other small molecules as intercellular pathogenic signals. FEMS Microbiol. Lett. 2011, 314, 10–17.
- Schimmel, T.G.; Coffman, A.D.; Parsons, S.J. Effect of Butyrolactone I on the Producing Fungus, Aspergillus terreus. Appl. Environ. Microbiol. 1998, 64, 3707–3712.
- Cottier, F.; Mühlschlegel, F.A. Communication in Fungi. Int. J. Microbiol. 2012, 2012, 351832.
- Albuquerque, P.; Casadevall, A. Quorum sensing in fungi—A review. Med. Mycol. 2012, 50, 337–345.
- Estrela, A.B.; Abraham, W.R. Fungal Metabolites for the Control of Biofilm Infections. Agriculture 2016, 6, 37.
- Padder, S.A.; Prasad, R.; Shah, A.H. Quorum sensing: A less known mode of communication among fungi. Microbiol. Res. 2018, 210, 51–58.
- Haas, H. Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Nat. Prod. Rep. 2014, 31, 1266–1276.
- Al Shaer, D.; Al Musaimi, O.; de la Torre, B.G.; Albericio, F. Hydroxamate siderophores: Natural occurrence, chemical synthesis, iron binding affinity and use as Trojan horses against pathogens. Eur. J. Med. Chem. 2020, 208, 112791.
- Rangel, L.I.; Hamilton, O.; de Jonge, R.; Bolton, M.D. Fungal social influencers: Secondary metabolites as a platform for shaping the plant-associated community. Plant J. 2021, 108, 632–645.
- Kosanić, M.; Ranković, B. Lichen Secondary Metabolites as Potential Antibiotic Agents. In Lichen Secondary Metabolites; Ranković, B., Ed.; Springer: Cham, Switzerland, 2019; pp. 99–127.
- Robey, M.T.; Caesar, L.K.; Drott, M.T.; Keller, N.P.; Kelleher, N.L. An interpreted atlas of biosynthetic gene clusters from 1,000 fungal genomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2020230118.
- Wei, B.; Du, A.Q.; Zhou, Z.Y.; Lai, C.; Yu, W.C.; Yu, J.B.; Yu, Y.L.; Chen, J.W.; Zhang, H.W.; Xu, X.W.; et al. An atlas of bacterial secondary metabolite biosynthesis gene clusters. Environ. Microbiol. 2021, 23, 6981–6992.
- Bharadwaj, R.; Kumar, S.R.; Sharma, A.; Sathishkumar, R. Plant Metabolic Gene Clusters: Evolution, Organization, and Their Applications in Synthetic Biology. Front. Plant Sci. 2021, 12, 697318.
- Alami, M.M.; Ouyang, Z.; Zhang, Y.; Shu, S.; Yang, G.; Mei, Z.; Wang, X. The Current Developments in Medicinal Plant Genomics Enabled the Diversification of Secondary Metabolites’ Biosynthesis. Int. J. Mol. Sci. 2022, 23, 15932.
- García-Estrada, C.; Domínguez-Santos, R.; Kosalková, K.; Martín, J.F. Transcription Factors Controlling Primary and Secondary Metabolism in Filamentous Fungi: The β-Lactam Paradigm. Fermentation 2018, 4, 47.
- Gluck-Thaler, E.; Haridas, S.; Binder, M.; Grigoriev, I.V.; Crous, P.W.; Spatafora, J.W.; Bushley, K.; Slot, J.C. The Architecture of Metabolism Maximizes Biosynthetic Diversity in the Largest Class of Fungi. Mol. Biol. Evol. 2020, 37, 2838–2856.
- Wang, W.; Yu, Y.; Keller, N.P.; Wang, P. Presence, Mode of Action, and Application of Pathway Specific Transcription Factors in Aspergillus Biosynthetic Gene Clusters. Int. J. Mol. Sci. 2021, 22, 8709.
- Almeida, H.; Palys, S.; Tsang, A.; Diallo, A.B. TOUCAN: A framework for fungal biosynthetic gene cluster discovery. NAR Genom. Bioinform. 2020, 2, lqaa098.
- Duban, M.; Cociancich, S.; Leclère, V. Nonribosomal Peptide Synthesis Definitely Working Out of the Rules. Microorganisms 2022, 10, 577.
- Cummings, M.; Breitling, R.; Takano, E. Steps towards the synthetic biology of polyketide biosynthesis. FEMS Microbiol. Lett. 2014, 351, 116–125.
- Cane, D.E.; Walsh, C.T. The parallel and convergent universes of polyketide synthases and nonribosomal peptide synthetases. Chem. Biol. 1999, 6, R319–R325.
- Ansari, M.Z.; Yadav, G.; Gokhale, R.S.; Mohanty, D. NRPS-PKS: A knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 2004, 32, W405–W413.
- Wang, H.; Fewer, D.P.; Holm, L.; Rouhiainen, L.; Sivonen, K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc. Natl. Acad. Sci. USA 2014, 111, 9259–9264.
- Du, L.; Sánchez, C.; Shen, B. Hybrid peptide-polyketide natural products: Biosynthesis and prospects toward engineering novel molecules. Metab. Eng. 2001, 3, 78–95.
- Miller, B.R.; Gulick, A.M. Structural Biology of Non-Ribosomal Peptide Synthetases. In Nonribosomal Peptide and Polyketide Biosynthesis. Methods in Molecular Biology; Evans, B., Ed.; Humana Press: New York, NY, USA, 2016; Volume 1401.
- Weber, T.; Marahiel, M.A. Exploring the Domain Structure of Modular Nonribosomal Peptide Synthetases. Structure 2001, 9, R3–R9.
- Challis, G.L.; Naismith, J.H. Structural aspects of non-ribosomal peptide biosynthesis. Curr. Opin. Struct. Biol. 2004, 14, 748–756.
- Stachelhaus, T.; Mootz, H.D.; Marahiel, M.A. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 1999, 6, 493–505.
- Bloudoff, K.; Schmeing, T.M. Structural and functional aspects of the nonribosomal peptide synthetase condensation domain superfamily: Discovery, dissection and diversity. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1587–1604.
- Von Döhren, H.; Dieckmann, R.; Pavela-Vrancic, M. The nonribosomal code. Chem. Biol. 1999, 6, R273–R279.
- Kalb, D.; Lackner, G.; Hoffmeister, D. Fungal peptide synthetases: An update on functions and specificity signatures. Fungal Biol. Rev. 2013, 27, 43–50.
- Walsh, C.T.; O’Brien, R.V.; Khosla, C. Nonproteinogenic Amino Acid Building Blocks for Nonribosomal Peptide and Hybrid Polyketide Scaffolds. Angew. Chem. Int. Ed. 2013, 52, 7098–7124.
- Fichtner, M.; Voigt, K.; Schuster, S. The tip and hidden part of the iceberg: Proteinogenic and non-proteinogenic aliphatic amino acids. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 3258–3269.
- Hubbard, B.K.; Thomas, M.G.; Walsh, C.T. Biosynthesis of L-p-hydroxyphenylglycine, a non-proteinogenic amino acid constituent of peptide antibiotics. Chem. Biol. 2000, 7, 931–942.
- Felnagle, E.A.; Jackson, E.E.; Chan, Y.A.; Podevels, A.M.; Berti, A.D.; McMahon, M.D.; Thomas, M.G. Nonribosomal Peptide Synthetases Involved in the Production of Medically Relevant Natural Products. Mol. Pharm. 2008, 5, 191–211.
- Crawford, A.; Wilson, D. Essential metals at the host-pathogen interface: Nutritional immunity and micronutrient assimilation by human fungal pathogens. FEMS Yeast Res. 2015, 15, fov071.
- Gerwien, F.; Skrahina, V.; Kasper, L.; Hube, B.; Brunke, S. Metals in fungal virulence. FEMS Microbiol. Rev. 2018, 42, fux050.
- Cheng, Y.Q.; Tang, G.L.; Shen, B. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc. Natl. Acad. Sci. USA 2003, 100, 3149–3154.
- Cheng, Y.Q.; Coughlin, J.M.; Lim, S.K.; Shen, B. Type I Polyketide Synthases That Require Discrete Acyltransferases. Methods Enzymol. 2009, 459, 165–186.
- Tang, G.L.; Cheng, Y.Q.; Shen, B. Leinamycin Biosynthesis Revealing Unprecedented Architectural Complexity for a Hybrid Polyketide Synthase and Nonribosomal Peptide Synthetase. Chem. Biol. 2004, 11, 33–45.
- Cox, R.J.; Skellam, E.; Williams, K. Biosynthesis of Fungal Polyketides. In Physiology and Genetics; Anke, T., Schüffler, A., Eds.; Springer: Cham, Switzerland, 2018; Volume 15, pp. 385–412.
- Hang, L.; Liu, N.; Tang, Y. Coordinated and Iterative Enzyme Catalysis in Fungal Polyketide Biosynthesis. ACS Catal. 2016, 6, 5935–5945.
- Cacho, R.A.; Thuss, J.; Xu, W.; Sanichar, R.; Gao, Z.; Nguyen, A.; Vederas, J.C.; Tang, Y. Understanding Programming of Fungal Iterative Polyketide Synthases: The Biochemical Basis for Regioselectivity by the Methyltransferase Domain in the Lovastatin Megasynthase. J. Am. Chem. Soc. 2015, 137, 15688–15691.
- Smith, J.L.; Skiniotis, G.; Sherman, D.H. Architecture of the polyketide synthase module: Surprises from electron cryo-microscopy. Curr. Opin. Struct. Biol. 2015, 31, 9–19.
- Navarro-Muñoz, J.C.; Collemare, J. Evolutionary Histories of Type III Polyketide Synthases in Fungi. Front. Microbiol. 2020, 10, 3018.
- Chooi, Y.H.; Tang, Y. Navigating the fungal polyketide chemical space: From genes to molecules. J. Org. Chem. 2012, 77, 9933–9953.
- Ames, B.D.; Nguyen, C.; Bruegger, J.; Smith, P.; Xu, W.; Ma, S.; Wong, E.; Wong, S. Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis. Proc. Natl. Acad. Sci. USA 2012, 109, 11144–11149.
- Vinha, A.F.; Machado, M.; Beatriz, M.; Oliveira, P.P. Terpenes from Fungi. In Natural Secondary Metabolites; Carocho, M., Heleno, S.A., Barros, L., Eds.; Springer: Cham, Switzerland, 2023; pp. 497–528.
- Bian, G.; Deng, Z.; Liu, T. Strategies for terpenoid overproduction and new terpenoid discovery. Curr. Opin. Biotechnol. 2017, 48, 234–241.
- Christianson, D.W. Structural and Chemical Biology of Terpenoid Cyclases. Chem. Rev. 2017, 117, 11570–11648.
- Pan, X.; Du, W.; Zhang, X.; Lin, X.; Li, F.R.; Yang, Q.; Wang, H.; Rudolf, J.D.; Zhang, B.; Dong, L. Bin Discovery, Structure, and Mechanism of a Class II Sesquiterpene Cyclase. J. Am. Chem. Soc. 2022, 144, 22067–22074.
- Gao, Y.; Honzatko, R.B.; Peters, R.J. Terpenoid synthase structures: A so far incomplete view of complex catalysis. Nat. Prod. Rep. 2012, 29, 1153–1175.
- Rudolf, J.D.; Chang, C.Y. Terpene synthases in disguise: Enzymology, structure, and opportunities of non-canonical terpene synthases. Nat. Prod. Rep. 2020, 37, 425–463.
- Buckingham, J.; Cooper, C.M.; Purchase, R. Natural Products Desk Reference; CRC Press: Boca Raton, FL, USA, 2015.
- Nes, W.D. Biosynthesis of cholesterol and other sterols. Chem. Rev. 2011, 111, 6423–6451.
- Minami, A.; Ugai, T.; Ozaki, T.; Oikawa, H. Predicting the chemical space of fungal polyketides by phylogeny-based bioinformatics analysis of polyketide synthase-nonribosomal peptide synthetase and its modification enzymes. Sci. Rep. 2020, 10, 13556.
- Silva, J.; Garcia, J.; Guimarães, R.; Palito, C.; Lemos, A.; Barros, L.; Alves, M.J.; Silva, J.; Garcia, J.; Guimarães, R.; et al. Alkaloids from Fungi. In Natural Secondary Metabolites; Carocho, M., Heleno, S.A., Barros, L., Eds.; Springer: Cham, Switzerland, 2023; pp. 529–554.
- Kishimoto, S.; Sato, M.; Tsunematsu, Y.; Watanabe, K. Evaluation of Biosynthetic Pathway and Engineered Biosynthesis of Alkaloids. Molecules 2016, 21, 1078.
- Gerhards, N.; Neubauer, L.; Tudzynski, P.; Li, S.M. Biosynthetic Pathways of Ergot Alkaloids. Toxins 2014, 6, 3281–3295.
- Mai, P.; Li, S.M. Alkaloids Derived from Tryptophan: A Focus on Ergot Alkaloids. In Natural Products: Phytochemistry, Botany and Metabolism of Alkaloids, Phenolics and Terpenes; Springer: Berlin/Heidelberg, Germany, 2013; pp. 683–714.
- Dai, G.; Shen, Q.; Zhang, Y.; Bian, X. Biosynthesis of Fungal Natural Products Involving Two Separate Pathway Crosstalk. J. Fungi 2022, 8, 320.
- Boettger, D.; Hertweck, C. Molecular diversity sculpted by fungal PKS-NRPS hybrids. Chembiochem 2013, 14, 28–42.
- Nickles, G.R.; Oestereicher, B.; Keller, N.P.; Drott, M.T. Mining for a New Class of Fungal Natural Products: The Evolution, Diversity, and Distribution of Isocyanide Synthase Biosynthetic Gene Clusters. bioRxiv 2023.
- Martín, J.F.; Álvarez-Álvarez, R.; Liras, P. Clavine Alkaloids Gene Clusters of Penicillium and Related Fungi: Evolutionary Combination of Prenyltransferases, Monooxygenases and Dioxygenases. Genes 2017, 8, 342.
- Raffa, N.; Won, T.H.; Sukowaty, A.; Candor, K.; Cui, C.; Halder, S.; Dai, M.; Landero-Figueroa, J.A.; Schroeder, F.C.; Keller, N.P. Dual-purpose isocyanides produced by Aspergillus fumigatus contribute to cellular copper sufficiency and exhibit antimicrobial activity. Proc. Natl. Acad. Sci. USA 2021, 118, e2015224118.
- Carroll, C.S.; Grieve, C.L.; Murugathasan, I.; Bennet, A.J.; Czekster, C.M.; Lui, H.; Naismith, J.; Moore, M.M. The rhizoferrin biosynthetic gene in the fungal pathogen Rhizopus delemar is a novel member of the NIS gene family. Int. J. Biochem. Cell Biol. 2017, 89, 136–146.
- Terabayashi, Y.; Sano, M.; Yamane, N.; Marui, J.; Tamano, K.; Sagara, J.; Dohmoto, M.; Oda, K.; Ohshima, E.; Tachibana, K.; et al. Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genet. Biol. 2010, 47, 953–961.
- Hicks, C.; Witte, T.E.; Sproule, A.; Lee, T.; Shoukouhi, P.; Popovic, Z.; Menzies, J.G.; Boddy, C.N.; Liu, M.; Overy, D.P. Evolution of the ergot alkaloid biosynthetic gene cluster results in divergent mycotoxin profiles in claviceps purpurea sclerotia. Toxins 2021, 13, 861.
- Chen, T.Y.; Chen, J.; Tang, Y.; Zhou, J.; Guo, Y.; Chang, W.C. Current Understanding toward Isonitrile Group Biosynthesis and Mechanism. Chin. J. Chem. 2021, 39, 463–472.
- Liu, S.; Widom, J.; Kemp, C.W.; Crews, C.M.; Clardy, J. Structure of human methionine aminopeptidase-2 complexed with fumagillin. Science 1998, 282, 1324–1327.
- Terfehr, D.; Dahlmann, T.A.; Kück, U. Transcriptome analysis of the two unrelated fungal β-lactam producers Acremonium chrysogenum and Penicillium chrysogenum: Velvet-regulated genes are major targets during conventional strain improvement programs. BMC Genom. 2017, 18, 272.
- Biermann, F.; Wenski, S.L.; Helfrich, E.J.N. Navigating and expanding the roadmap of natural product genome mining tools. Beilstein J. Org. Chem. 2022, 18, 1656–1671.
- Kwon, M.J.; Steiniger, C.; Cairns, T.C.; Wisecaver, J.H.; Lind, A.; Pohl, C.; Regner, C.; Rokas, A.; Meyer, V. Beyond the biosynthetic gene cluster paradigm: Genome-wide co-expression networks connect clustered and unclustered transcription factors to secondary metabolic pathways. bioRxiv 2020.
- Sélem-Mojica, N.; Aguilar, C.; Gutiérrez-García, K.; Martínez-Guerrero, C.E.; Barona-Gómez, F. EvoMining reveals the origin and fate of natural product biosynthetic enzymes. Microb. Genom. 2019, 5, e000260.
- Schüller, A.; Studt-Reinhold, L.; Strauss, J. How to Completely Squeeze a Fungus—Advanced Genome Mining Tools for Novel Bioactive Substances. Pharmaceutics 2022, 14, 1837.
- Gutiérrez, S.A.; Velasco, J.A.; Fernandez, F.J.; Martín, J.F. The cefG gene of Cephalosporium acremonium is linked to the cefEF gene and encodes a deacetylcephalosporin C acetyltransferase closely related to homoserine O-acetyltransferase. J. Bacteriol. 1992, 174, 3056–3064.
- Dumina, M.V.; Zhgun, A.A.; Domracheva, A.G.; Novak, M.I.; El’darov, M.A. Chromosomal polymorphism of Acremonium chrysogenum strains producing cephalosporin C. Russ. J. Genet. 2012, 48, 778–784.
- Walsh, C.T.; Chen, H.; Keating, T.A.; Hubbard, B.K.; Losey, H.C.; Luo, L.; Marshall, C.G.; Miller, D.A.; Patel, H.M. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr. Opin. Chem. Biol. 2001, 5, 525–534.
- Little, R.F.; Hertweck, C. Chain release mechanisms in polyketide and non-ribosomal peptide biosynthesis. Nat. Prod. Rep. 2022, 39, 163–205.
- Xu, W.; Chooi, Y.-H.; Choi, J.W.; Li, S.; Vederas, J.C.; Da Silva, N.A.; Tang, Y. LovG: The Thioesterase Required for Dihydromonacolin L Release and Lovastatin Nonaketide Synthase Turnover in Lovastatin Biosynthesis. Angew. Chemie Int. Ed. 2013, 52, 6472–6475.
- Martín, J.F. Transport systems, intracellular traffic of intermediates and secretion of β-lactam antibiotics in fungi. Fungal Biol. Biotechnol. 2020, 7, 6.
- Ullán, R.V.; Casqueiro, J.; Bañuelos, O.; Fernández, F.J.; Gutiérrez, S.; Martín, J.F. A novel epimerization system in fungal secondary metabolism involved in the conversion of isopenicillin N into penicillin N in Acremonium chrysogenum. J. Biol. Chem. 2002, 277, 46216–46225.
- Zhgun, A.; Dumina, M.; Valiakhmetov, A.; Eldarov, M. The critical role of plasma membrane H+-ATPase activity in cephalosporin C biosynthesis of Acremonium chrysogenum. PLoS ONE 2020, 15, e0238452.
- Durairaj, P.; Hur, J.S.; Yun, H. Versatile biocatalysis of fungal cytochrome P450 monooxygenases. Microb. Cell Fact. 2016, 15, 125.
- Lu, W.; Feng, J.; Chen, X.; Bao, Y.J.; Wang, Y.; Wu, Q.; Ma, Y.; Zhu, D. Distinct regioselectivity of fungal P450 enzymes for steroidal hydroxylation. Appl. Environ. Microbiol. 2019, 85, e01182-19.
- Van Den Brink, H.M.; Van Gorcom, R.F.M.; Van Den Hondel, C.A.M.J.J.; Punt, P.J. Cytochrome P450 Enzyme Systems in Fungi. Fungal Genet. Biol. 1998, 23, 1–17.
- Zhgun, A.; El’darov, M.; Solodar’, L.; Sokolov, N.; Archakov, A.; Skriabin, K. Heterologous expression of eukaryotic CYP450. 1. Heterologous expression of cytochrome P450 2B4 using groups with various affinity in E. coli. Vopr. Meditsinskoi Khimii 2001, 47, 382–392.
- Zhang, X.; Guo, J.; Cheng, F.; Li, S. Cytochrome P450 enzymes in fungal natural product biosynthesis. Nat. Prod. Rep. 2021, 38, 1072–1099.
- Barriuso, J.; Nguyen, D.T.; Li, J.W.-H.; Roberts, J.N.; MacNevin, G.; Chaytor, J.L.; Marcus, S.L.; Vederas, J.C.; Ro, D.-K. Double Oxidation of the Cyclic Nonaketide Dihydromonacolin L to Monacolin J by a Single Cytochrome P450 Monooxygenase, LovA. J. Am. Chem. Soc. 2011, 133, 8078–8081.
- Kennedy, J.; Auclair, K.; Kendrew, S.G.; Park, C.; Vederas, J.C.; Hutchinson, C.R. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science 1999, 284, 1368–1372.
- Lah, L.; Podobnik, B.; Novak, M.; Korošec, B.; Berne, S.; Vogelsang, M.; Kraševec, N.; Zupanec, N.; Stojan, J.; Bohlmann, J.; et al. The versatility of the fungal cytochrome P450 monooxygenase system is instrumental in xenobiotic detoxification. Mol. Microbiol. 2011, 81, 1374–1389.
- Sang, H.; Hulvey, J.P.; Green, R.; Xu, H.; Im, J.; Chang, T.; Jung, G. A Xenobiotic Detoxification Pathway through Transcriptional Regulation in Filamentous Fungi. mBio 2018, 9, 457–475.
- Shin, J.; Kim, J.E.; Lee, Y.W.; Son, H. Fungal Cytochrome P450s and the P450 Complement (CYPome) of Fusarium graminearum. Toxins 2018, 10, 112.
- Chen, W.; Lee, M.K.; Jefcoate, C.; Kim, S.C.; Chen, F.; Yu, J.H. Fungal Cytochrome P450 Monooxygenases: Their Distribution, Structure, Functions, Family Expansion, and Evolutionary Origin. Genome Biol. Evol. 2014, 6, 1620–1634.
- Crits-Christoph, A.; Bhattacharya, N.; Olm, M.R.; Song, Y.S.; Banfield, J.F. Transporter genes in biosynthetic gene clusters predict metabolite characteristics and siderophore activity. Genome Res. 2021, 31, 239–250.
- Fernández-Aguado, M.; Martín, J.F.; Rodríguez-Castro, R.; García-Estrada, C.; Albillos, S.M.; Teijeira, F.; Ullán, R. V New insights into the isopenicillin N transport in Penicillium chrysogenum. Metab. Eng. 2014, 22, 89–103.
- Zhgun, A.A.; Nuraeva, G.K.; Eldarov, M.A. The Role of LaeA and LovE Regulators in Lovastatin Biosynthesis with Exogenous Polyamines in Aspergillus terreus. Appl. Biochem. Microbiol. 2019, 55, 639–648.
- Yang, J.; Xu, X.; Liu, G. Amplification of an MFS Transporter Encoding Gene penT Significantly Stimulates Penicillin Production and Enhances the Sensitivity of Penicillium chrysogenum to Phenylacetic Acid. J. Genet. Genom. 2012, 39, 593–602.
- Ramzan, R.; Virk, M.S.; Chen, F. The ABCT31 Transporter Regulates the Export System of Phenylacetic Acid as a Side-Chain Precursor of Penicillin G in Monascus ruber M7. Front. Microbiol. 2022, 13, 2054.
- Zhgun, A.A.; Eldarov, M.A. Spermidine and 1,3-Diaminopropane Have Opposite Effects on the Final Stage of Cephalosporin C Biosynthesis in High-Yielding Acremonium chrysogenum Strain. Int. J. Mol. Sci. 2022, 23, 14625.
- Ullán, R.V.; Teijeira, F.; Guerra, S.M.; Vaca, I.; Martín, J.F. Characterization of a novel peroxisome membrane protein essential for conversion of isopenicillin N into cephalosporin C. Biochem. J. 2010, 432, 227–236.
- Teijeira, F.; Ullán, R.V.; Guerra, S.M.; García-Estrada, C.; Vaca, I.; Martín, J.F. The transporter CefM involved in translocation of biosynthetic intermediates is essential for cephalosporin production. Biochem. J. 2009, 418, 113–124.
- Yan, Y.; Liu, N.; Tang, Y. Recent developments in self-resistance gene directed natural product discovery. Nat. Prod. Rep. 2020, 37, 879–892.
- Dumina, M.V.; Zhgun, A.A.; Kerpichnikov, I.V.; Domracheva, A.G.; Novak, M.I.; Valiachmetov, A.Y.; Knorre, D.A.; Severin, F.F.; Eldarov, M.A.; Bartoshevich, Y.E. Functional analysis of MFS protein CefT involved in the transport of beta-lactam antibiotics in Acremonium chrysogenum and Saccharomyces cerevisiae. Appl. Biochem. Microbiol. 2013, 49, 368–377.
- McLean, T.C.; Wilkinson, B.; Hutchings, M.I.; Devine, R. Dissolution of the Disparate: Co-ordinate Regulation in Antibiotic Biosynthesis. Antibiotics 2019, 8, 83.
- Teijeira, F.; Ullán, R.V.; Fernández-Aguado, M.; Martín, J.F. CefR modulates transporters of beta-lactam intermediates preventing the loss of penicillins to the broth and increases cephalosporin production in Acremonium chrysogenum. Metab. Eng. 2011, 13, 532–543.
More
