A bottom-up route towards predicting evolution relies on a deep understanding of the complex network that proteins form inside cells. In a rapidly expanding panorama of experimental possibilities, the most difficult question is how to conceptually approach the disentangling of such complex networks. These can exhibit varying degrees of hierarchy and modularity, which obfuscate protein functions that may prove pivotal for adaptation. Using the well-established polarity network in budding yeast as a case study, we organize current literature to highlight protein entrenchments inside polarity in five sub modules: timing, mating, bud-scar, reaction-diffusion and the actin pathway.
- Yeast Cell Polarity
- Evolution
- Cdc42
- Redundancy
- Reaction-diffusion
- Actin
- Bud-scar
- Mating
- Modularity
- Neutrality
Thanks so much for your check. We sincerely hope you may create this entry, since you are the expert in this academic research. You can click the “submit” button to upload it and revise it. We will help you layout after you submit it. Moreover, we will link your article at the entry, and more scholars and students can look through it.
1. Introduction
How cells work and how they evolve is at the heart of cell biology. In this work we will review how cellular architecture (“how cells work”) and its evolutionary properties (“how they evolve”) are related to each other. Understanding evolution and possible mutational paths of protein networks, and especially the cell polarity network, is not only satisfying our curiosity but may also help us understand and possibly predict cancer progression[1].
Every cell consists of many different interconnected functional protein networks (for definitions, see
), such as transcription, translation, or polarity establishment[2]. The network’s architecture, (for example: which protein binds to/reacts with which other protein), impacts the evolutionary possibilities of a network in multiple ways. For example, hubs, proteins with many binding partners, tend to evolve slower[3]. Less connected proteins, that may be deleted in a cell without a detectable change in cell physiology, can permit duplication of other genes and thus promote evolution[4]: duplicates of a gene enable new options for diversification, which facilitate further evolution of a gene/protein and the surrounding network[5][6][7]. Interestingly, many mutations (from 3% of non-silent mutations in bacteria to 30% in hominids[8]) in a cell show very weak, or no effect on the cell’s function, a phenomenon called neutrality[9]. Thus, proteins that may seem unimportant for how the cell works now, in this environment, may become important when changes occur in the network architecture due to a mutation or a switch in environment[10][11].
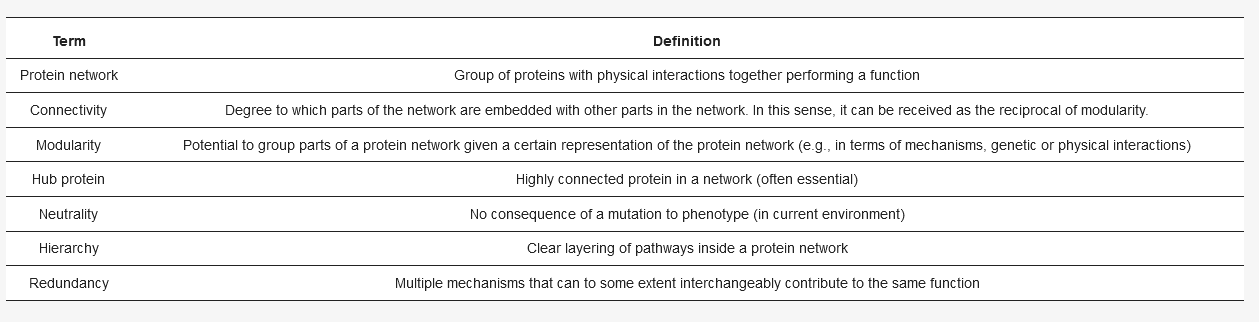
Definitions of important terms used throughout the introduction.
A well-studied model organism to concretize how these proteins, without a detectable phenotype, shape a network is
, or budding yeast. The organism generally exhibits many of the network properties defined in
, such as hierarchy and the presence of hubs[2]. Here, neutrality is also pervasive, as only 40% of homozygous gene deletions for the entire organism initially had obvious phenotypes[12]. Moreover, the environment has been shown to have a notable influence on neutrality, as lethal heterozygous deletions can be compensated by poor medium[13]. As a general rule, both the network architecture and the environment can mask the function of many proteins.
Within this organism, a, to some extent, representative example of a protein network is the polarity network, which governs how the yeast chooses a direction in which to divide and involves directing dozens of proteins in a process of breaking its internal spherical symmetry (see, e.g.,[14]). As required, we observe the presence of the common network properties demonstrated in
, such as hierarchy and redundancy. Polarity is also one of many biological functions in yeast for which a subdivision of many proteins into a quasi-modular network proved possible[15]. Within polarity, even more detailed submodules can be distinguished[16]. Neutrality is also exhibited by several polarity proteins, which is in part responsible for the difficulty in determining the role of each protein. Lastly, polarity is a pattern formation process where, by definition, spatiotemporal dependencies are important, and understanding evolution generally relies on understanding this type of dependencies[17].
However, the polarity network is not a prime example of the sort of networks with abundant transcriptional regulation. Other templates are better suited for learning about the evolution of gene regulation, such as interaction networks centered around transcriptional regulators (e.g., Mcm1[18]), ribosomal regulation (e.g.,[19]) the stress response (e.g.,[20]) or metabolic response (e.g.,
pathway[21]). The existence of established regulatory templates thus conveniently complements our focus on symmetry breaking during polarity as a model for the protein interaction network, which is also a topic for evolutionary studies.
Concretely, in[22] a mutant strongly defective in polarity establishment was experimentally evolved and found to recover remarkably reproducibly, e.g., the first rescuing mutation to sweep the population was always the same. Because of this exhibited tractability of the adaptations, network structures within the polarity network that facilitated evolution could be concretely interpreted in terms of redundancies[23]. In another approach to determine the flexibility of the polarity network, historical evolution was studied for 40+ proteins in almost 300 fungal species in[11]. Again, the polarity network exhibited sufficient modularity so that studying its evolution separately from other functions still yielded interpretable results. For example, authors showed that polarity network size was shaped in part by fungal lifestyle (e.g., uni- or multicellular).
Nevertheless, clear justification for observed evolutionary trajectories in this network remain difficult to make. It is sometimes possible to generate more abstract predictions by linking network architecture to the evolvability of the polarity network using classical regulatory motifs. For example, the presence of positive and negative feedback in polarity establishment confers robustness to the network[24][25], which in turn may facilitate evolution. But in order to make more concrete and detailed predictions about evolutionary trajectories, an important insight is that we need mechanistic information of (parts of) the polarity network.
To illustrate this point, a bottom-up model was constructed in[26] where molecular details were coarse-grained following analysis on numerical simulations of multiple polarity mechanisms[23]. This approach was successful in quantitatively describing the fitness along the evolutionary trajectory in[22]. Furthermore, the predictions on epistasis, an important bottleneck for predicting evolution[27], can be extended to other modules, and although use of full mechanistic understanding is superior in quantitative assessments of epistasis, biofunctional information (viz. from GO-terms, in agreement with[28]) as input to the model suffices for epistasis sign predictions.
Instructive is the Nrp1 case where full information is absent, but some phenomenological information is available. Based on the latter, inclusion of Nrp1 into the bottom-up coarse-grained model of[26] is still worthwhile, but leaves room for improvement. This marks the importance of continuing to investigate protein networks until molecular mechanisms have been elucidated. In this review we advocate that obtaining this knowledge is (soon) feasible, motivating the use of the yeast polarity system for studies in network organization (with properties such as hierarchy, modularity/connectivity and redundancy that can “hide” proteins) as well as evolution.
2. Polarity Overview
Within the yeast polarity network, four pathways to polarization exist which cannot easily be considered modular. Their interconnectivity can be conveniently visualized in the form of a Venn diagram (
). These pathways are hierarchically set up, in the following order: the mating pathway at the top, then the bud scar pathway, followed by the reaction−diffusion pathway and finally the actin pathway. In short, their function boils down to the act of condensing the GTPase Cdc42 bound to GTP molecules (i.e., active Cdc42) to one point on the plasma membrane, which can signal downstream effectors to proceed the cell cycle[29]. To prevent premature or overdue localization of active Cdc42 and allow some influence on the hierarchy of pathways, a fifth pathway exists to control the previous four, namely the timing pathway. The next section summarizes the most important interactions in and across all pathways, starting with the timing cue. As a site note, we have done our best to include all relevant papers, but apologize for important papers we have missed.
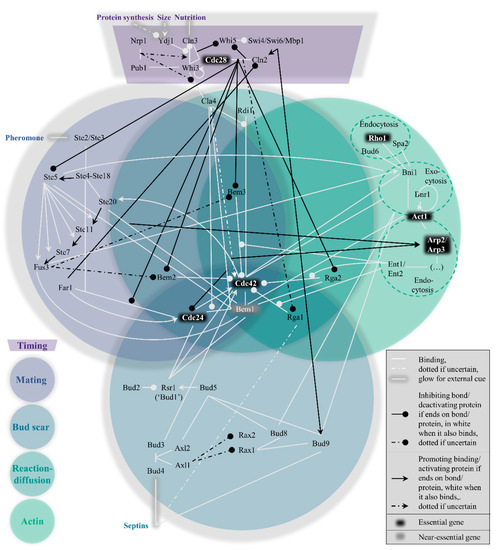
Venn diagram representation of the hierarchical, nonmodular pathway structure for protein−protein interactions in budding yeast polarity establishment (
for references).
2.1. Timing: The Control Knob
During isotropic growth in G1, active Cdc42 localization is suppressed by overactivity of its associated GTPase activating proteins (GAPs) and sequestration of its guanine nucleotide exchange factor (GEF). The consequence of both circumstances is the vast abundance of inactive Cdc42, which is bound to GDP instead of GTP[30], rendering it impossible to signal the polarity cue. The purpose of this pathway (see top dark purple region in
) is hence to timely reduce GAP activity and release the GEF, which must be in response to important physiological parameters that indicate the readiness of the cell: sufficient protein production, a sufficient size, and sufficient nutrition.
The physiological state of the cell enters the equation through nuclear levels of cyclin Cln3. Upon sufficient nutrition and size, Cln3 levels rise either more directly through higher Cln3 mRNA abundance[31], or more indirectly through Ydj1 disturbing Cln3 localization by Whi3[32], the latter also being an inhibitor of Cln3 mRNA translation[33]. The arrival of nuclear Cln3 allows binding partner and cyclin-dependent kinase Cdc28[34] to phosphorylate Whi5, which had inhibited expression of Cln2, another cyclin[35][36]. Cln2 can then reinforce its own expression, consolidating the original Cln3 signal[37].
Now, the Cdc28-Cln2 complex can distribute the physiological signal to the aforementioned targets, the GAPs and the GEF. The kinase Cdc28 phosphorylates all four GAPs Bem2, Bem3, Rga1 and Rga2[38][39][40][41] and Far1, which was keeping the GEF Cdc24 in the nucleus[42]. Now cytoplasmic levels of active Cdc42 can rise, leading to polarity establishment through subsequent pathways.
Importantly, the completion of the timing pathway causes the hierarchy of the subsequent pathways to change. While the mating pathway is otherwise dominant, the kinase Cdc28 phosphorylates Ste5, a crucial hub in the mating pathway, to stop the mating in its tracks[43][44][45]. In the following discussion of the mating pathway, the situation is considered where the timing pathway did not overwrite its behavior.
2.2. Mating: Heavily Cross-Linked
The mating pathway is the dominant force across the four symmetry-breaking pathways. While polarization in a random orientation is possible after the timing cue (see the section on reaction−diffusion further on), the presence of pheromones of the opposite mating type (a or α) should redirect the Cdc42 localization to the side of the pheromone signal. This process revolves around Ste5, as also depicted in the left, blue-grey circle of
.
Briefly put, once pheromones bind membrane proteins Ste2 and Ste3[46][47], Ste4 is released from the membrane[48][49] and binds Ste20 and scaffold Ste5. This scaffold binds Ste7, Ste11 and Fus3, which are activated by sequential phosphorylation[50][51][52]. Fus3 may inhibit the GAPs Bem2 and Bem3[39], while Ste5 binds the GEF Cdc24[53], replacing the absence of the timing pathway result to stimulate activity of Cdc42.
While this simplified view would suffice to redirect the Cdc42 localization, the mating pathway is much more intertwined with the other pathways than seemingly necessary, particularly with the actin pathway. The abundant mechanistic redundancies result in a more complex picture, obfuscating the role of the proteins involved. For example, active Cdc42 stimulates Ste11 phosphorylation/activation[54]. Another form of positive feedback, as well as a bridge to the actin pathway, is the Cdc42 recruitment of formin Bni1[55]. The resulting nucleation of actin cables may transport Ste5-GEF Cdc24 complexes[56], possibly also through Bem1. This scaffold co-immunoprecipitates with Act1[57] and Far1[58], which is bound to the GEF Cdc24[59], but is itself in turn also bound to Bem1[60]. Another actin cross-link is the phosphorylation and localization of Bni1 through Fus3[61]. Clearly, care must be taken in assigning roles to different proteins, as many are overloaded.
2.3. Bud Scar: Mostly Modular and Ordered
In the absence of a mating cue, the timing pathway reduces GAP activity and releases the GEF, while the mating pathway is repressed. Under the new hierarchy, the bud scar pathway is normally dominant. The scar refers to leftover proteins from the previous division, named septins[62][63]. This spatial cue can be exploited for polarity establishment; a new bud forms adjacent to the scar (axial budding, haploids) or also at the opposing side (bipolar budding, diploids)[64][65]. The bottom, dark blue circle of
represents this path from septins to Cdc42 recruitment graphically.
An important bud scar localization target is Bud5, which activates and recruits Bud1[66][67], a protein also known as Rsr1[65][68]. In haploids, where Axl1 is specifically expressed[69], the Bud5 localization occurs by relay of the septin signal to a protein complex of Bud3, Bud4, Axl1, Axl2 and Bud5[70][71][72][73]. In diploids, functionality of Rax1 and Rax2 is not impaired, presumably by blocked expression of Axl1[74], so these can localize Bud9 and Bud8 to the bud scar or the opposite end of the scar respectively[75], which in turn recruit Bud5[76].
After Bud5, localization follows of Bud2[77], the GAP for Bud1[78], to complete the control of the GTPase cycle of Bud1. Finally, Bud1 binds GEF Cdc24 and Bem1[79] (although Cdc24 has the strongest affinity with Bem1[80]), to redirect the pattern formation made possible after the timing cue. As linkage of Cdc42 GAP Rga1 to septins prevents reuse of the previous location[81], the new bud forms adjacent to the bud scar.
As a whole, the bud scar pathway is not completely modular either. Aside from nudging the reaction−diffusion pathway (see next section), an example of a cross-link is that Bud8 and Bud9 are delivered by actin transport[82]. The highest position in the hierarchy in the absence of the mating cue is also not absolute; multiple ways to promote the subsequent reaction−diffusion pathway exist, such as deletion of Bud1 and Bud8[83], Axl2 and Rax1 [84], or Bem1[22].
2.4. Reaction−Diffusion: Ample Redundancy
Even in the absence of chemical or spatial cues, the shift in balance towards activation of Cdc42 induced by the timing pathway still provides the conditions for swift symmetry breaking. Theoretical models concerning this pathway have been subject to updates as more molecular details have been revealed. Central is the strong positive feedback generated by the Bem1-GEF Cdc24 complex, as modelled in, e.g.,[85][86] with further refinement in[87]. This feedback is sufficient for polarity success, which becomes rather insensitive to GAP abundance. More details on the GAPs were uncovered in[23].
What makes this pathway special is the limited number of proteins that are unique to this pathway, as seen from the central, emerald circle in
, namely only Cla4 and Rdi1. The latter is the least cross-linked of the two, providing a possible justification for referring to the WT mechanism as the Rdi1 polarity mechanism, as in[24]. Cla4 is more context-dependent, possibly having two opposing roles, promoting and inhibiting polarity[23][88][89]. Yet both Rdi1 and Cla4 are dispensable for polarity[88][90].
An even stronger addition to the redundancy within this pathway is on the positive feedback side. Without Bem1, generic rescuing feedbacks suffice [
], among which Cla4 could account for 20% of their function[91]. More feedbacks may be found in the GAPs through actin transport as described in, e.g., [
]. This brings us to the actin pathway as the final layer to discuss.
2.5. Actin: The Mysterious Auxiliary Layer
The actin pathway (rightmost green circle in
) has featured several times already in the previously discussed pathways, but its individual role is still quite uncertain. Yeast formin Bni1 which nucleates actin cables, binds active Cdc42[55], and is known to be involved in exo- and endocytosis[92][93]. This suggests transport of polarity proteins from and to the presumptive bud site. The resulting actin pathway has been confusingly implicated in two opposing roles; promoting Cdc42 polarization, see, e.g.,[24][94][95], as well as negatively impacting Cdc42 polarization[96][97].
A way to reconcile these findings is that actin transport contributes to a process promoting Cdc42 polarization but without relying on significant transport of Cdc42 itself. As mentioned in the mating pathway, Bem1 and Act1 co-immunoprecipitate[57], suggesting that Bem1, and concordantly its multiple binding partners, might get transported through the actin pathway. However, in the absence of Bem1, 80% of the positive feedback is still unidentified[91], which may very well be actin-related. Instead, a prime candidate is the GAP group, which is known to bind the epsin-coating of actin cables involved in endocytosis[98]. In any case, it is quite difficult to decipher the actin pathway, in large part due to its low positioning in the yeast polarity hierarchy.
References
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306–313, doi:10.1038/nature10762.
- Costanzo, M.; VanderSluis, B.; Koch, E.N.; Baryshnikova, A.; Pons, C.; Tan, G.; Wang, W.; Usaj, M.; Hanchard, J.; Lee, S.D. A global genetic interaction network maps a wiring diagram of cellular function. Science 2016, 353, doi:10.1126/science.aaf1420.
- Fraser, H.B.; Wall, D.P.; Hirsh, A.E. A simple dependence between protein evolution rate and the number of protein-protein interactions. BMC Evol. Biol. 2003, 3, 1–6.
- Ratmann, O.; Wiuf, C.; Pinney, J.W. From evidence to inference: Probing the evolution of protein interaction networks. HFSP J. 2009, 3, 290–306.
- Zhang, J. Evolution by gene duplication: An update. Trends Ecol. Evol. 2003, 18, 292–298, doi:10.1016/S0169-5347(03)00033-8.
- Näsvall, J.; Sun, L.; Roth, J.R.; Andersson, D.I. Real-time evolution of new genes by innovation, amplification, and divergence. Science 2012, 338, 384–387.
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161.
- Eyre-Walker, A.; Keightley, P.D. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 2007, 8, 610–618, doi:10.1038/nrg2146.
- Thoday, J.M. Non-Darwinian “evolution” and biological progress. Nature 1975, 255, 675–677.
- Khan, A.I.; Dinh, D.M.; Schneider, D.; Lenski, R.E.; Cooper, T.F. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 2011, 332, 1193–1196.
- Diepeveen, E.T.; Gehrmann, T.; Pourquié, V.; Abeel, T.; Laan, L. Patterns of Conservation and Diversification in the Fungal Polarization Network. Genome Biol. Evol. 2018, 10, 1765–1782, doi:10.1093/gbe/evy121.
- Winzeler, E.A. Functional Characterization of the S. cerevisiae Genome by Gene Deletion and Parallel Analysis. Science 1999, 285, 901–906.
- Deutschbauer, A.M.; Jaramillo, D.F.; Proctor, M.; Kumm, J.; Hillenmeyer, M.E.; Davis, R.W.; Nislow, C.; Giaever, G. Mechanisms of Haploinsufficiency Revealed by Genome-Wide Profiling in Yeast. Genetics 2005, 169, 1915–1925, doi:10.1534/genetics.104.036871.
- Martin, S.G.; Arkowitz, R.A. Cell polarization in budding and fission yeasts. FEMS Microbiol. Rev. 2014, 38, 228–253, doi:10.1111/1574-6976.12055.
- Costanzo, M.; Baryshnikova, A.; Bellay, J.; Kim, Y.; Spear, E.D.; Sevier, C.S.; Ding, H.; Koh, J.L.; Toufighi, K.; Mostafavi, S.; et al. The genetic landscape of a cell. Science 2010, 327, 425–431.
- Gao, J.T.; Guimera, R.; Li, H.; Pinto, I.M.; Sales-Pardo, M.; Wai, S.C.; Rubinstein, B.; Li, R. Modular coherence of protein dynamics in yeast cell polarity system. Proc. Natl. Acad. Sci. USA 2011, 108, 7647–7652, doi:10.1073/pnas.1017567108.
- Yamada, T.; Bork, P. Evolution of biomolecular networks—Lessons from metabolic and protein interactions. Nat. Rev. Mol. Cell Biol. 2009, 10, 791–803, doi:10.1038/nrm2787.
- Tuch, B.B.; Galgoczy, D.J.; Hernday, A.D.; Li, H.; Johnson, A.D. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 2008, 6, e38.
- Tanay, A.; Regev, A.; Shamir, R. Conservation and evolvability in regulatory networks: The evolution of ribosomal regulation in yeast. Proc. Natl. Acad. Sci. USA 2005, 102, 7203–7208.
- Dhar, R.; Sagesser, R.; Weikert, C.; Wagner, A. Yeast Adapts to a Changing Stressful Environment by Evolving Cross-Protection and Anticipatory Gene Regulation. Mol. Biol. Evol. 2013, 30, 573–588, doi:10.1093/molbev/mss253.
- Slot, J.C.; Rokas, A. Multiple GAL pathway gene clusters evolved independently and by different mechanisms in fungi. Proc. Natl. Acad. Sci. USA 2010, 107, 10136–10141, doi:10.1073/pnas.0914418107.
- Laan, L.; Koschwanez, J.H.; Murray, A.W. Evolutionary adaptation after crippling cell polarization follows reproducible trajectories. eLife 2015, 4, e09638.
- Brauns, F.; de la Cruz, L.M.I.; Daalman, W.K.-G.; de Bruin, I.; Halatek, J.; Laan, L.; Frey, E. Adaptability and evolution of the cell polarization machinery in budding yeast. bioRxiv 2020, doi:10.1101/2020.09.09.290510.
- Freisinger, T.; Klünder, B.; Johnson, J.; Müller, N.; Pichler, G.; Beck, G.; Costanzo, M.; Boone, C.; Cerione, R.A.; Frey, E.; et al. Establishment of a robust single axis of cell polarity by coupling multiple positive feedback loops. Nat. Commun. 2013, 4, 1807, doi:10.1038/ncomms2795.
- Howell, A.S.; Jin, M.; Wu, C.-F.; Zyla, T.R.; Elston, T.C.; Lew, D.J. Negative Feedback Enhances Robustness in the Yeast Polarity Establishment Circuit. Cell 2012, 149, 322–333, doi:10.1016/j.cell.2012.03.012.
- Daalman, W.K.-G.; Laan, L. Predicting an epistasis-rich genotype-phenotype map with a coarse-grained bottom-up model of budding yeast polarity. bioRxiv 2020, doi:10.1101/2020.11.09.374363.
- Breen, M.S.; Kemena, C.; Vlasov, P.K.; Notredame, C.; Kondrashov, F.A. Epistasis as the primary factor in molecular evolution. Nature 2012, 490, 535–538, doi:10.1038/nature11510.
- Yu, M.K.; Kramer, M.; Dutkowski, J.; Srivas, R.; Licon, K.; Kreisberg, J.F.; Ng, C.T.; Krogan, N.; Sharan, R.; Ideker, T. Translation of Genotype to Phenotype by a Hierarchy of Cell Subsystems. Cell Syst. 2016, 2, 77–88, doi:10.1016/j.cels.2016.02.003.
- Park, H.-O.; Bi, E. Central Roles of Small GTPases in the Development of Cell Polarity in Yeast and Beyond. Microbiol. Mol. Biol. Rev. 2007, 71, 48–96, doi:10.1128/MMBR.00028-06.
- Hall, A. The cellular functions of small GTP-binding proteins. Science 1990, 249, 635–640.
- Gallego, C.; Garí, E.; Colomina, N.; Herrero, E.; Aldea, M. The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J. 1997, 16, 7196–7206.
- Vergés, E.; Colomina, N.; Garí, E.; Gallego, C.; Aldea, M. Cyclin Cln3 Is Retained at the ER and Released by the J Chaperone Ydj1 in Late G1 to Trigger Cell Cycle Entry. Mol. Cell 2007, 26, 649–662, doi:10.1016/j.molcel.2007.04.023.
- Cai, Y.; Futcher, B. Effects of the Yeast RNA-Binding Protein Whi3 on the Half-Life and Abundance of CLN3 mRNA and Other Targets. PLoS ONE 2013, 8, e84630, doi:10.1371/journal.pone.0084630.
- Wang, H.; Garí, E.; Vergés, E.; Gallego, C.; Aldea, M. Recruitment of Cdc28 by Whi3 restricts nuclear accumulation of the G1 cyclin–Cdk complex to late G1. EMBO J. 2004, 23, 180–190.
- Koch, C.; Schleiffer, A.; Ammerer, G.; Nasmyth, K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 1996, 10, 129–141.
- Costanzo, M.; Nishikawa, J.L.; Tang, X.; Millman, J.S.; Schub, O.; Breitkreuz, K.; Dewar, D.; Rupes, I.; Andrews, B.; Tyers, M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 2004, 117, 899–913.
- Stuart, D.; Wittenberg, C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995, 9, 2780–2794.
- Archambault, V.; Chang, E.J.; Drapkin, B.J.; Cross, F.R.; Chait, B.T.; Rout, M.P. Targeted proteomic study of the cyclin-Cdk module. Mol. Cell 2004, 14, 699–711.
- Knaus, M.; Pelli-Gulli, M.-P.; Van Drogen, F.; Springer, S.; Jaquenoud, M.; Peter, M. Phosphorylation of Bem2p and Bem3p may contribute to local activation of Cdc42p at bud emergence. EMBO J. 2007, 26, 4501–4513.
- McCusker, D.; Denison, C.; Anderson, S.; Egelhofer, T.A.; Yates, J.R.; Gygi, S.P.; Kellogg, D.R. Cdk1 coordinates cell-surface growth with the cell cycle. Nat. Cell Biol. 2007, 9, 506–515, doi:10.1038/ncb1568.
- Sopko, R.; Huang, D.; Smith, J.C.; Figeys, D.; Andrews, B.J. Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J. 2007, 26, 4487–4500.
- Shimada, Y.; Gulli, M.-P.; Peter, M. Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating. Nat. Cell Biol. 2000, 2, 117–124.
- Garrenton, L.S.; Braunwarth, A.; Irniger, S.; Hurt, E.; Kunzler, M.; Thorner, J. Nucleus-Specific and Cell Cycle-Regulated Degradation of Mitogen-Activated Protein Kinase Scaffold Protein Ste5 Contributes to the Control of Signaling Competence. Mol. Cell. Biol. 2009, 29, 582–601, doi:10.1128/MCB.01019-08.
- Doncic, A.; Falleur-Fettig, M.; Skotheim, J.M. Distinct Interactions Select and Maintain a Specific Cell Fate. Mol. Cell 2011, 43, 528–539, doi:10.1016/j.molcel.2011.06.025.
- Doncic, A.; Skotheim, J.M. Feedforward Regulation Ensures Stability and Rapid Reversibility of a Cellular State. Mol. Cell 2013, 50, 856–868, doi:10.1016/j.molcel.2013.04.014.
- Blumer, K.J.; Reneke, J.E.; Thorner, J. The STE2 gene product is the ligand-binding component of the alpha-factor receptor of Saccharomyces cerevisiae. J. Biol. Chem. 1988, 263, 10836–10842.
- Hagen, D.C.; McCaffrey, G.; Sprague, G.F. Evidence the yeast STE3 gene encodes a receptor for the peptide pheromone a factor: Gene sequence and implications for the structure of the presumed receptor. Proc. Natl. Acad. Sci. USA 1986, 83, 1418–1422.
- Whiteway, M.; Hougan, L.; Dignard, D.; Thomas, D.Y.; Bell, L.; Saari, G.C.; Grant, F.J.; O’Hara, P.; MacKay, V.L. The STE4 and STE18 genes of yeast encode potential β and γ subunits of the mating factor receptor-coupled G protein. Cell 1989, 56, 467–477.
- Hirschman, J.E.; De Zutter, G.S.; Simonds, W.F.; Jenness, D.D. The Gβγ Complex of the Yeast Pheromone Response Pathway SUBCELLULAR FRACTIONATION AND PROTEIN-PROTEIN INTERACTIONS. J. Biol. Chem. 1997, 272, 240–248.
- Chol, K.-Y.; Satterberg, B.; Lyons, D.M.; Elion, E.A. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell 1994, 78, 499–512.
- Feng, Y.; Song, L.Y.; Kincaid, E.; Mahanty, S.K.; Elion, E.A. Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr. Biol. 1998, 8, 267–282.
- Good, M.; Tang, G.; Singleton, J.; Reményi, A.; Lim, W.A. The Ste5 Scaffold Directs Mating Signaling by Catalytically Unlocking the Fus3 MAP Kinase for Activation. Cell 2009, 136, 1085–1097, doi:10.1016/j.cell.2009.01.049.
- Wang, Y.; Chen, W.; Simpson, D.M.; Elion, E.A. Cdc24 Regulates Nuclear Shuttling and Recruitment of the Ste5 Scaffold to a Heterotrimeric G Protein in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 13084–13096, doi:10.1074/jbc.M410461200.
- Lamson, R.E.; Winters, M.J.; Pryciak, P.M. Cdc42 Regulation of Kinase Activity and Signaling by the Yeast p21-Activated Kinase Ste20. Mol. Cell. Biol. 2002, 22, 2939–2951, doi:10.1128/MCB.22.9.2939-2951.2002.
- Evangelista, M.; Blundell, K.; Longtine, M.S.; Chow, C.J.; Adames, N.; Pringle, J.R.; Peter, M.; Boone, C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 1997, 276, 118–122.
- Qi, M.; Elion, E.A. Formin-induced actin cables are required for polarized recruitment of the Ste5 scaffold and high level activation of MAPK Fus3. J. Cell Sci. 2005, 118, 2837–2848, doi:10.1242/jcs.02418.
- Leeuw, T.; Fourest-Lieuvin, A.; Wu, C.; Chenevert, J.; Clark, K.; Whiteway, M.; Thomas, D.Y.; Leberer, E. Pheromone Response in Yeast: Association of Bem1p with Proteins of the MAP Kinase Cascade and Actin. Science 1995, 1210–1213.
- Lyons, D.M.; Mahanty, S.K.; Choi, K.-Y.; Manandhar, M.; Elion, E.A. The SH3-domain protein Bem1 coordinates mitogen-activated protein kinase cascade activation with cell cycle control in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996, 16, 4095–4106.
- Butty, A.-C.; Pryciak, P.M.; Huang, L.S.; Herskowitz, I.; Peter, M. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 1998, 282, 1511–1516.
- Peterson, J. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho- type GTPases in yeast. J. Cell Biol. 1994, 127, 1395–1406, doi:10.1083/jcb.127.5.1395.
- Matheos, D.; Metodiev, M.; Muller, E.; Stone, D.; Rose, M.D. Pheromone-induced polarization is dependent on the Fus3p MAPK acting through the formin Bni1p. J. Cell Biol. 2004, 165, 99–109, doi:10.1083/jcb.200309089.
- Kim, H.B. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: Localization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol. 1991, 112, 535–544, doi:10.1083/jcb.112.4.535.
- Flescher, E.G. Components required for cytokinesis are important for bud site selection in yeast. J. Cell Biol. 1993, 122, 373–386, doi:10.1083/jcb.122.2.373.
- Freifelder, D. Bud position in Saccharomyces cerevisiae. J. Bacteriol. 1960, 80, 567.
- Chant, J.; Herskowitz, I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 1991, 65, 1203–1212.
- Chant, J.; Corrado, K.; Pringle, J.R.; Herskowitz, I. Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1. Cell 1991, 65, 1213–1224.
- Kang, P.J.; Béven, L.; Hariharan, S.; Park, H.-O. The Rsr1/Bud1 GTPase interacts with itself and the Cdc42 GTPase during bud-site selection and polarity establishment in budding yeast. Mol. Biol. Cell 2010, 21, 3007–3016.
- Bender, A.; Pringle, J.R. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc. Natl. Acad. Sci. USA 1989, 86, 9976–9980.
- Fujita, A.; Oka, C.; Arikawa, Y.; Katagai, T.; Tonouchi, A.; Kuhara, S.; Misumi, Y. A yeast gene necessary for bud-site selection encodes a protein similar to insulin-degrading enzymes. Nature 1994, 372, 567.
- Chant, J.; Mischke, M.; Mitchell, E.; Herskowitz, I.; Pringle, J.R. Role of Bud3p in producing the axial budding pattern of yeast. J. Cell Biol. 1995, 129, 767–778.
- Sanders, S.L.; Herskowitz, I. The BUD4 protein of yeast, required for axial budding, is localized to the mother/BUD neck in a cell cycle-dependent manner. J. Cell Biol. 1996, 134, 413–427.
- Kang, P.J.; Angerman, E.; Jung, C.-H.; Park, H.-O. Bud4 mediates the cell-type-specific assembly of the axial landmark in budding yeast. J. Cell Sci. 2012, 125, 3840–3849, doi:10.1242/jcs.103697.
- Kang, P.J.; Hood-DeGrenier, J.K.; Park, H.-O. Coupling of septins to the axial landmark by Bud4 in budding yeast. J. Cell Sci. 2013, 126, 1218–1226, doi:10.1242/jcs.118521.
- Bi, E.; Park, H.-O. Cell Polarization and Cytokinesis in Budding Yeast. Genetics 2012, 191, 347–387, doi:10.1534/genetics.111.132886.
- Kang, P.J.; Angerman, E.; Nakashima, K.; Pringle, J.R.; Park, H.-O. Interactions among Rax1p, Rax2p, Bud8p, and Bud9p in marking cortical sites for bipolar bud-site selection in yeast. Mol. Biol. Cell 2004, 15, 5145–5157.
- Krappmann, A.-B.; Taheri, N.; Heinrich, M.; Mösch, H.-U. Distinct domains of yeast cortical tag proteins Bud8p and Bud9p confer polar localization and functionality. Mol. Biol. Cell 2007, 18, 3323–3339.
- Marston, A.L.; Chen, T.; Yang, M.C.; Belhumeur, P.; Chant, J. A localized GTPase exchange factor, Bud5, determines the orientation of division axes in yeast. Curr. Biol. 2001, 11, 803–807.
- Bender, A. Genetic evidence for the roles of the bud-site-selection genes BUD5 and BUD2 in control of the Rsr1p (Bud1p) GTPase in yeast. Proc. Natl. Acad. Sci. USA 1993, 90, 9926–9929.
- Park, H.-O.; Bi, E.; Pringle, J.R.; Herskowitz, I. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc. Natl. Acad. Sci. USA 1997, 94, 4463–4468.
- Zheng, Y.; Bender, A.; Cerione, R.A. Interactions among proteins involved in bud-site selection and bud-site assembly in Saccharomyces cerevisiae. J. Biol. Chem. 1995, 270, 626–630.
- Tong, Z.; Gao, X.-D.; Howell, A.S.; Bose, I.; Lew, D.J.; Bi, E. Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein mediated zone of inhibition. J. Cell Biol. 2007, 179, 1375–1384, doi:10.1083/jcb.200705160.
- Schenkman, L.R.; Caruso, C.; Pagé, N.; Pringle, J.R. The role of cell cycle–regulated expression in the localization of spatial landmark proteins in yeast. J. Cell Biol. 2002, 156, 829–841, doi:10.1083/jcb.200107041.
- Irazoqui, J.E.; Gladfelter, A.S.; Lew, D.J. Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 2003, 5, 1062–1070, doi:10.1038/ncb1068.
- Smith, S.E.; Rubinstein, B.; Mendes Pinto, I.; Slaughter, B.D.; Unruh, J.R.; Li, R. Independence of symmetry breaking on Bem1-mediated autocatalytic activation of Cdc42. J. Cell Biol. 2013, 202, 1091–1106, doi:10.1083/jcb.201304180.
- Ozbudak, E.M.; Becskei, A.; van Oudenaarden, A. A System of Counteracting Feedback Loops Regulates Cdc42p Activity during Spontaneous Cell Polarization. Dev. Cell 2005, 9, 565–571, doi:10.1016/j.devcel.2005.08.014.
- Goryachev, A.B.; Pokhilko, A.V. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 2008, 582, 1437–1443, doi:10.1016/j.febslet.2008.03.029.
- Klünder, B.; Freisinger, T.; Wedlich-Söldner, R.; Frey, E. GDI-Mediated Cell Polarization in Yeast Provides Precise Spatial and Temporal Control of Cdc42 Signaling. PLoS Comput. Biol. 2013, 9, e1003396, doi:10.1371/journal.pcbi.1003396.
- Tiedje, C.; Sakwa, I.; Just, U.; Höfken, T. The rho gdi rdi1 regulates rho gtpases by distinct mechanisms. Mol. Biol. Cell 2008, 19, 2885–2896.
- Kuo, C.-C.; Savage, N.S.; Chen, H.; Wu, C.-F.; Zyla, T.R.; Lew, D.J. Inhibitory GEF Phosphorylation Provides Negative Feedback in the Yeast Polarity Circuit. Curr. Biol. 2014, 24, 753–759, doi:10.1016/j.cub.2014.02.024.
- Cvrckova, F.; De Virgilio, C.; Manser, E.; Pringle, J.R.; Nasmyth, K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995, 9, 1817–1830.
- Daalman, W.K.-G. Coupling of Genotype-Phenotype Maps to Noise-Driven Adaptation, Showcased in Yeast Polarity; TU Delft: Delft, the Netherlands, 2020,doi:10.4233/uuid:0dec7916-fa0c-44ef-85ef-0694d3eb25b3.
- Pruyne, D.; Gao, L.; Bi, E.; Bretscher, A. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell 2004, 15, 4971–4989.
- Prosser, D.C.; Drivas, T.G.; Maldonado-Báez, L.; Wendland, B. Existence of a novel clathrin-independent endocytic pathway in yeast that depends on Rho1 and formin. J. Cell Biol. 2011, 195, 657–671, doi:10.1083/jcb.201104045.
- Wedlich-Soldner, R.; Altschuler, S.; Wu, L.; Li, R. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 2003, 299, 1231–1235.
- Slaughter, B.D.; Unruh, J.R.; Das, A.; Smith, S.E.; Rubinstein, B.; Li, R. Non-uniform membrane diffusion enables steady-state cell polarization via vesicular trafficking. Nat. Commun. 2013, 4, 1380, doi:10.1038/ncomms2370.
- Layton, A.T.; Savage, N.S.; Howell, A.S.; Carroll, S.Y.; Drubin, D.G.; Lew, D.J. Modeling Vesicle Traffic Reveals Unexpected Consequences for Cdc42p-Mediated Polarity Establishment. Curr. Biol. 2011, 21, 184–194, doi:10.1016/j.cub.2011.01.012.
- Watson, L.J.; Rossi, G.; Brennwald, P. Quantitative Analysis of Membrane Trafficking in Regulation of Cdc42 Polarity. Traffic 2014, 15, 1330–1343, doi:10.1111/tra.12211.
- Aguilar, R.C.; Longhi, S.A.; Shaw, J.D.; Yeh, L.-Y.; Kim, S.; Schön, A.; Freire, E.; Hsu, A.; McCormick, W.K.; Watson, H.A.; et al. Epsin N-terminal homology domains perform an essential function regulating Cdc42 through binding Cdc42 GTPase-activating proteins. PNAS 2006, 103, 4116–4121.
