Biobanks provide a platform for innovative biomedical research. Recent scientific advances in cryopreservation have enabled the prospect of establishing “living biobanks” that store viable, functional tissue or replicable cell types for years to decades.
- patient-derived cancer models
- living biobanks
1. Introduction
Traditional cancer models including cell lines and animal models have limited applications in both basic and clinical cancer research. Genomics-based precision oncology only help 2–20% patients with solid cancer. Functional diagnostics and patient-derived cancer models are needed for precision cancer biology. Researchers will summarize applications of conditional cell reprogramming (CR) in cancer research and next generation living biobanks (NGLB). Together with organoids, CR has been cited in two NCI (National Cancer Institute, USA) programs (PDMR: patient-derived cancer model repository; HCMI: human cancer model initiatives. HCMI will be distributed through ATCC). Briefly, the CR method is a simple co-culture technology with a Rho kinase inhibitor, Y-27632, in combination with fibroblast feeder cells, which allows researchersto rapidly expand both normal and malignant epithelial cells from diverse anatomic sites and mammalian species and does not require transfection with exogenous viral or cellular genes. Establishment of CR cells from both normal and tumor tissue is highly efficient. The robust nature of the technique is exemplified by the ability to produce 2 × 106 cells in five days from a core biopsy of tumor tissue. Normal CR cell cultures retain a normal karyotype and differentiation potential and CR cells derived from tumors retain their tumorigenic phenotype. CR also allows researchers to enrich cancer cells from urine (for bladder cancer), blood (for prostate cancer), and pleural effusion (for non-small cell lung carcinoma). The ability to produce inexhaustible cell populations using CR technology from small biopsies and cryopreserved specimens has the potential to transform biobanking repositories (NGLB: next-generation living biobank) and current pathology practice by enabling genetic, biochemical, metabolomic, proteomic, and biological assays, including chemosensitivity testing as a functional diagnostics tool for precision cancer medicine.
2. Patient-Derived Cancer Models and Cell Cultures are Needed for Precision Oncology
2.1. iPS (Induce Pluripotent Stem) Cells
2.2. Organoid Cultures
2.3. Patient-Derived Xenografts (PDX)
2.4. Conditionally Reprogrammed Cells (CRCs)
3. Next-Generation Living Biobanks (NGLB)
Living Biobanks will have a significant impact across basic biological research, medicine and the biopharma industry; however, the effects of such applications are underexplored. Recent approaches, for example, patient-derived xenograft models, organoids, conditionally reprogrammed cells, induced pluripotent cells, and other cancer precision medicine applications, represent an unexhausted resource of living biobanks. However, these concepts are applied in very many different ways by the academic and private sectors, representing an actively growing field that has yet to reach clinical consensus or maturity. Here researchers summarize application of CR technology in living biobanks. 28–29 March 2011, researchers first presented “bring biobank to life” at BRN 2011 Symposium “Advancing Cancer Research Through Biospecimen Science” of NCI when researchers started CR technology [26][40]. Editor in Chief of American Journal of Pathology proposed applications of CR technology in living biobanks [40]. Eventually, more and more articles came out for tumor living biobanks as the applications of CRC and organoid cultures [13][19][22][28][36][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58]. However, that will be impossible task in terms of efforts, work load and cost if one would like to establish living biobanks from every patient in the hospital for the immediate use of personalized medicine. In order to balance of distribution of disease types for research and industry, it’s extremely important to cryopreserved tissue specimens and will generate cell cultures using CR and/or organoids technologies. Thus, researchers brought forth a new concept: next generation living biobank (NGLB) (Figure 61). Except for immediate use of personalized diagnostics and treatment, researchers suggest to collect and cryopreserve specimens including surgical specimens, core biopsies, needle biopsies, brushed cells, or cells from liquid biopsies (blood, urine, etc.). These specimens should be collected and stored together with their corresponding -omics (genomic, transcriptomic, proteomic, metabolomic, etc.) and clinical information. Then, CR technology will be used to generate an unexhausted cell cultures for living cell banks for future demands in drug discovery, disease modeling, regenerative medicine. These cryopreserved specimens, their corresponding CR cells and CRC- derivatives together compose of NGLB (Figure 1). Compared to regular living biobanks, this NGLB strategy would be a highly efficient platform with the minimal time, money, and other support, this may also avoid redundant cell lines from common diseases, for example, lung cancers. Most importantly, this NBLG includes cryopreserved specimens with broader coverage of disease types, which allow researchers to generate cell cultures from rare disease using CR or organoids cultures, since there are no available cell models or very few cell lines for many rare diseases, for example, adenoid cystic carcinoma (ACC), neuroendocrine cancer, etc. Researchers also list properties of conventional cell lines, CRC, organoids and PDX in basic and translational cancer research (Table 1).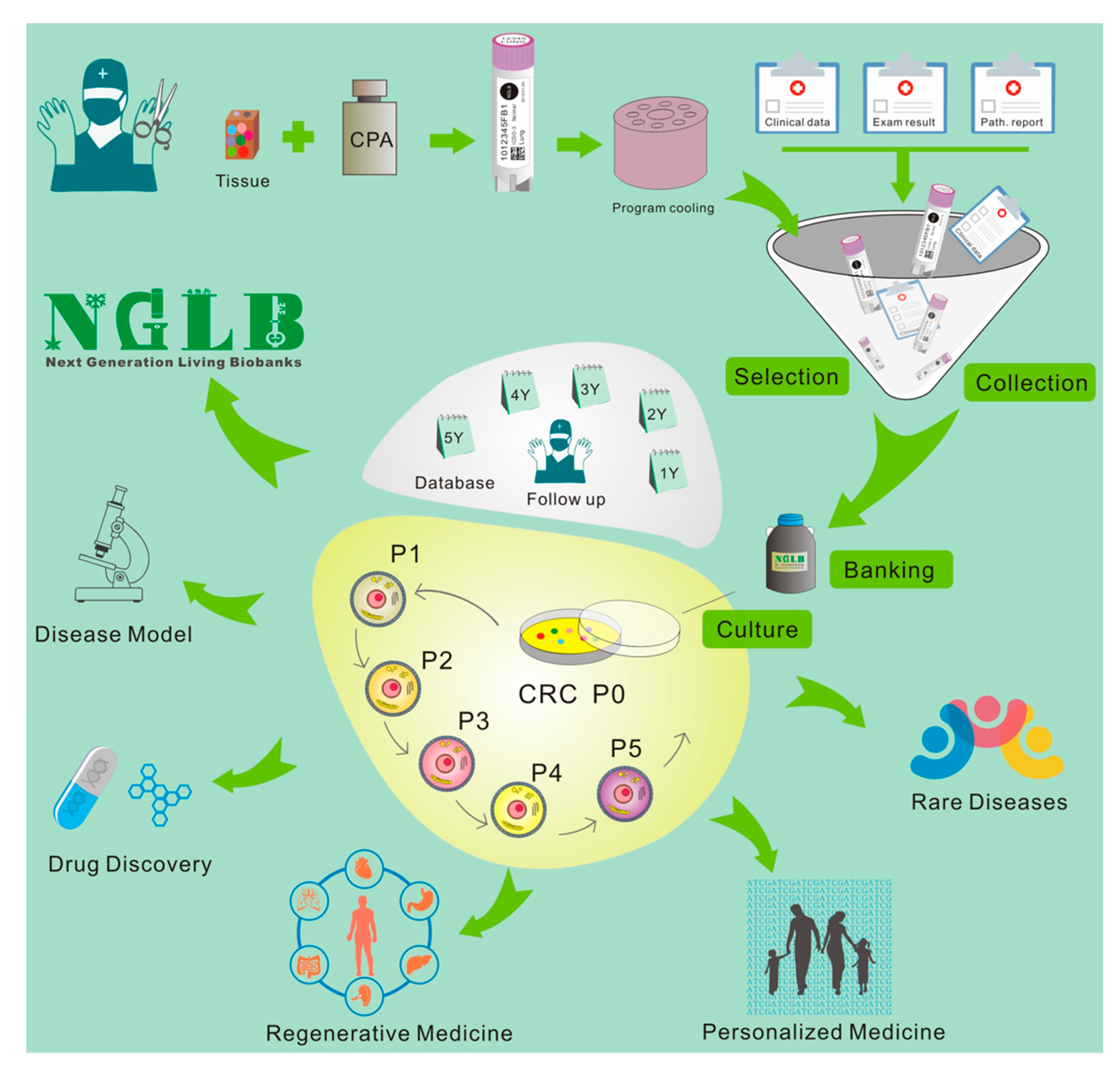
| Conventional Cell Lines | Organoids | PDX | CRC | |
|---|---|---|---|---|
| Sample size | ||||
| FNA | − | +/− | − | +++ |
| Core Biopsy | − | + | − | +++ |
| Surgical Specimens | + | +++ | ++ | +++ |
| Timing | dozen days | 1-5 weeks | 1-5mont | 1-10 days |
| Success rate of initiation | + 0–10% | ++ (5–80%) | ++ (2–30%) | +++50–100% |
| Tumor type specific | ||||
| Rapid Expansion | +++ | ++ | + | +++ |
| Matched Normal con | − | + | - | + |
| Karyotypic stability | − | ++ | N/A | ++ |
| 3D growth | − | + | +++ | − |
| Representation of tumor | + | ++ | ++ | ++ |
| Genetic manipulation | +++ | ++ | − | ++ |
| Maintenance (passage) | +++ | ++ | + | +++ |
| LT drug screens | +++ | – | + | +++ |
| HT drug screens | +++ | ++ | − | +++ |
| Heterogeneity | − | ++ | +++ | ++ |
| Tumor–stroma interaction | − | − | ++ | − |
References
- Curry, E.L.; Moad, M.; Robson, C.N.; Heer, R. Using induced pluripotent stem cells as a tool for modelling carcinogenesis. World J. Stem Cells 2015, 7, 461–469.
- Seki, T.; Fukuda, K. Methods of induced pluripotent stem cells for clinical application. World J. Stem Cells 2015, 7, 116–125.
- Marin Navarro, A.; Susanto, E.; Falk, A.; Wilhelm, M. Modeling cancer using patient-derived induced pluripotent stem cells to understand development of childhood malignancies. Cell Death Discov. 2018, 4, 7.
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced pluripotent stem cell technology: A decade of progress. Nat. Reviews. Drug Discov. 2017, 16, 115–130.
- Rowe, R.G.; Daley, G.Q. Induced pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Genet. 2019, 20, 377–388.
- Papaetrou, E.P. Author Correction: Patient-derived induced pluripotent stem cells in cancer research and precision oncology. Nat. Med. 2019, 25, 861.
- Papapetrou, E.P. Patient-derived induced pluripotent stem cells in cancer research and precision oncology. Nat. Med. 2016, 22, 1392–1401.
- Boj, S.F.; Hwang, C.I.; Baker, L.A.; Chio, I.I.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S.; et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015, 160, 324–338.
- Drost, J.; van Jaarsveld, R.H.; Ponsioen, B.; Zimberlin, C.; van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015, 521, 43–47.
- Li, X.; Nadauld, L.; Ootani, A.; Corney, D.C.; Pai, R.K.; Gevaert, O.; Cantrell, M.A.; Rack, P.G.; Neal, J.T.; Chan, C.W.; et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 2014, 20, 769–777.
- Nadauld, L.D.; Garcia, S.; Natsoulis, G.; Bell, J.M.; Miotke, L.; Hopmans, E.S.; Xu, H.; Pai, R.K.; Palm, C.; Regan, J.F.; et al. Metastatic tumor evolution and organoid modeling implicate TGFBR2 as a cancer driver in diffuse gastric cancer. Genome Biol. 2014, 15, 428.
- Sachs, N.; Clevers, H. Organoid cultures for the analysis of cancer phenotypes. Curr. Opin. Genet. Dev. 2014, 24, 68–73.
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945.
- Cheung, K.J.; Gabrielson, E.; Werb, Z.; Ewald, A.J. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell 2013, 155, 1639–1651.
- Ewald, A.J. 3D cell biology—the expanding frontier. J. Cell Sci. 2017, 130, 1.
- Shamir, E.R.; Ewald, A.J. Three-dimensional organotypic culture: Experimental models of mammalian biology and disease. Nat. Reviews. Mol. Cell Biol. 2014, 15, 647–664.
- Xu, H.; Lyu, X.; Yi, M.; Zhao, W.; Song, Y.; Wu, K. Organoid technology and applications in cancer research. J. Hematol. Oncol. 2018, 11, 116.
- Tuveson, D.; Clevers, H. Cancer modeling meets human organoid technology. Science 2019, 364, 952–955.
- Artegiani, B.; Clevers, H. Use and application of 3D-organoid technology. Hum. Mol. Genet. 2018, 27, R99–R107.
- Pauli, C.; Hopkins, B.D.; Prandi, D.; Shaw, R.; Fedrizzi, T.; Sboner, A.; Sailer, V.; Augello, M.; Puca, L.; Rosati, R.; et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov. 2017, 7, 462–477.
- Puca, L.; Bareja, R.; Prandi, D.; Shaw, R.; Benelli, M.; Karthaus, W.R.; Hess, J.; Sigouros, M.; Donoghue, A.; Kossai, M.; et al. Patient derived organoids to model rare prostate cancer phenotypes. Nat. Commun. 2018, 9, 2404.
- Xinaris, C. Organoids for replacement therapy: Expectations, limitations and reality. Curr. Opin. Organ. Transpl. 2019, 24, 555–561.
- Jin, K.; Teng, L.; Shen, Y.; He, K.; Xu, Z.; Li, G. Patient-derived human tumour tissue xenografts in immunodeficient mice: A systematic review. Clin. Transl. Oncol.: Off. Publ. Fed. Span. Oncol. Soc. Natl. Cancer Inst. Mex. 2010, 12, 473–480.
- Tentler, J.J.; Tan, A.C.; Weekes, C.D.; Jimeno, A.; Leong, S.; Pitts, T.M.; Arcaroli, J.J.; Messersmith, W.A.; Eckhardt, S.G. Patient-derived tumour xenografts as models for oncology drug development. Nat. Reviews. Clin. Oncol. 2012, 9, 338–350.
- Williams, S.A.; Anderson, W.C.; Santaguida, M.T.; Dylla, S.J. Patient-derived xenografts, the cancer stem cell paradigm, and cancer pathobiology in the 21st century. Lab. Investig. 2013, 93, 970–982.
- Liu, X.; Ory, V.; Chapman, S.; Yuan, H.; Albanese, C.; Kallakury, B.; Timofeeva, O.A.; Nealon, C.; Dakic, A.; Simic, V.; et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012, 180, 599–607.
- Suprynowicz, F.A.; Upadhyay, G.; Krawczyk, E.; Kramer, S.C.; Hebert, J.D.; Liu, X.; Yuan, H.; Cheluvaraju, C.; Clapp, P.W.; Boucher, R.C., Jr.; et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc. Natl. Acad. Sci. USA. 2012, 109, 20035–20040.
- Liu, X.; Krawczyk, E.; Suprynowicz, F.A.; Palechor-Ceron, N.; Yuan, H.; Dakic, A.; Simic, V.; Zheng, Y.-L.; Sripadhan, P.; Chen, C.; et al. Conditional reprogramming and long-term expansion of normal and tumor cells from human biospecimens. Nat. Protoc. 2017, 12, 439.
- Alamri, A.M.; Kang, K.; Groeneveld, S.; Wang, W.; Zhong, X.; Kallakury, B.; Hennighausen, L.; Liu, X.; Furth, P.A. Primary cancer cell culture: Mammary-optimized vs conditional reprogramming. Endocr. Relat. Cancer 2016, 23, 535–554.
- Gardell, A.M.; Qin, Q.; Rice, R.H.; Li, J.; Kültz, D. Derivation and osmotolerance characterization of three immortalized tilapia (Oreochromis mossambicus) cell lines. PLoS ONE 2014, 9, e95919.
- Wang, L.; Ye, L.; Wei, G.; Chen, Y.; Ye, L.; Wu, X.; Zeng, Z.; Wang, Y.; Yin, G.; Long, X.; et al. Conditional reprogrammed human limbal epithelial cells represent a novel in vitro cell model for drug responses. Biochem. Biophys. Res. Commun. 2018, 499, 735–742.
- Alkhilaiwi, F.; Wang, L.; Zhou, D.; Raudsepp, T.; Ghosh, S.; Paul, S.; Palechor-Ceron, N.; Brandt, S.; Luff, J.; Liu, X.; et al. Long-term expansion of primary equine keratinocytes that maintain the ability to differentiate into stratified epidermis. Stem Cell Res. Ther. 2018, 9, 181.
- O’Malley, Y.; Rotti, P.G.; Thornell, I.M.; Vanegas Calderón, O.G.; Febres-Aldana, C.; Durham, K.; Yao, J.; Li, X.; Zhu, Z.; Norris, A.W.; et al. Development of a polarized pancreatic ductular cell epithelium for physiological studies. J. Appl. Physiol. 2018, 125, 97–106.
- Saffari, P.S.; Vapniarsky, N.; Pollack, A.S.; Gong, X.; Vennam, S.; Pollack, A.J.; Verstraete, F.J.M.; West, R.B.; Arzi, B.; Pollack, J.R. Most canine ameloblastomas harbor HRAS mutations, providing a novel large-animal model of RAS-driven cancer. Oncogenesis 2019, 8, 11.
- McAuliffe, P.F.; Evans, K.W.; Akcakanat, A.; Chen, K.; Zheng, X.; Zhao, H.; Eterovic, A.K.; Sangai, T.; Holder, A.M.; Sharma, C.; et al. Ability to Generate Patient-Derived Breast Cancer Xenografts Is Enhanced in Chemoresistant Disease and Predicts Poor Patient Outcomes. PLoS ONE 2015, 10, e0136851.
- Saeed, K.; Rahkama, V.; Eldfors, S.; Bychkov, D.; Mpindi, J.P.; Yadav, B.; Paavolainen, L.; Aittokallio, T.; Heckman, C.; Wennerberg, K.; et al. Comprehensive Drug Testing of Patient-derived Conditionally Reprogrammed Cells from Castration-resistant Prostate Cancer. Eur. Urol. 2016.
- Beglyarova, N.; Banina, E.; Zhou, Y.; Mukhamadeeva, R.; Andrianov, G.; Bobrov, E.; Lysenko, E.; Skobeleva, N.; Gabitova, L.; Restifo, D.; et al. Screening of Conditionally Reprogrammed Patient-Derived Carcinoma Cells Identifies ERCC3-MYC Interactions as a Target in Pancreatic Cancer. Clin. Cancer Res. 2016, 22, 6153–6163.
- Walters, B.J.; Diao, S.; Zheng, F.; Walters, B.J.; Layman, W.S.; Zuo, J. Pseudo-immortalization of postnatal cochlear progenitor cells yields a scalable cell line capable of transcriptionally regulating mature hair cell genes. Sci. Rep. 2015, 5, 17792.
- Brown, D.D.; Dabbs, D.J.; Lee, A.V.; McGuire, K.P.; Ahrendt, G.M.; Bhargava, R.; Davidson, N.E.; Brufsky, A.M.; Johnson, R.R.; Oesterreich, S.; et al. Developing in vitro models of human ductal carcinoma in situ from primary tissue explants. Breast Cancer Res. Treat. 2015, 153, 311–321.
- Lisanti, M.P.; Tanowitz, H.B. Translational discoveries, personalized medicine, and living biobanks of the future. Am. J. Pathol. 2012, 180, 1334–1336.
- Sachs, N.; de Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H.; et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 2018, 172, 373–386 e310.
- Mullenders, J.; de Jongh, E.; Brousali, A.; Roosen, M.; Blom, J.P.A.; Begthel, H.; Korving, J.; Jonges, T.; Kranenburg, O.; Meijer, R.; et al. Mouse and human urothelial cancer organoids: A tool for bladder cancer research. Proc. Natl. Acad. Sci. USA 2019.
- Weeber, F.; Ooft, S.N.; Dijkstra, K.K.; Voest, E.E. Tumor Organoids as a Pre-clinical Cancer Model for Drug Discovery. Cell Chem. Biol. 2017.
- Mondal, A.M.; Ma, A.H.; Li, G.; Krawczyk, E.; Yuan, R.; Lu, J.; Schlegel, R.; Stamatakis, L.; Kowalczyk, K.J.; Philips, G.K.; et al. Fidelity of a PDX-CR model for bladder cancer. Biochem. Biophys. Res. Commun. 2019, 517, 49–56.
- Martini, A.; Sfakianos, J.P.; Galsky, M.D. Conditionally Reprogrammed Patient-derived Cells: A Step Forward Towards Personalized Medicine? Eur. Urol. 2019, 76, 435–436.
- Krawczyk, E.; Hong, S.H.; Galli, S.; Trinh, E.; Wietlisbach, L.; Misiukiewicz, S.F.; Tilan, J.U.; Chen, Y.S.; Schlegel, R.; Kitlinska, J. Murine neuroblastoma cell lines developed by conditional reprogramming preserve heterogeneous phenotypes observed in vivo. Lab. Investig. A J. Tech. Methods Pathol. 2019.
- Kettunen, K.; Bostrom, P.J.; Lamminen, T.; Heinosalo, T.; West, G.; Saarinen, I.; Kaipio, K.; Rantala, J.; Albanese, C.; Poutanen, M.; et al. Personalized Drug Sensitivity Screening for Bladder Cancer Using Conditionally Reprogrammed Patient-derived Cells. Eur. Urol. 2019, 76, 430–434.
- Jiang, S.; Wang, J.; Yang, C.; Tan, R.; Hou, J.; Shi, Y.; Zhang, H.; Ma, S.; Wang, J.; Zhang, M.; et al. Continuous culture of urine-derived bladder cancer cells for precision medicine. Protein Cell 2019.
- Yip, Y.L.; Lin, W.; Deng, W.; Jia, L.; Lo, K.W.; Busson, P.; Verillaud, B.; Liu, X.; Tsang, C.M.; Lung, M.L.; et al. Establishment of a nasopharyngeal carcinoma cell line capable of undergoing lytic Epstein-Barr virus reactivation. Lab. Investig. 2018, 98, 1093–1104.
- Saito, R.; Smith, C.C.; Utsumi, T.; Bixby, L.M.; Kardos, J.; Wobker, S.E.; Stewart, K.G.; Chai, S.; Manocha, U.; Byrd, K.M.; et al. Molecular Subtype-Specific Immunocompetent Models of High-Grade Urothelial Carcinoma Reveal Differential Neoantigen Expression and Response to Immunotherapy. Cancer Res. 2018, 78, 3954–3968.
- Jin, L.; Qu, Y.; Gomez, L.J.; Chung, S.; Han, B.; Gao, B.; Yue, Y.; Gong, Y.; Liu, X.; Amersi, F.; et al. Characterization of primary human mammary epithelial cells isolated and propagated by conditional reprogrammed cell culture. Oncotarget 2018, 9, 11503–11514.
- Alamri, A.M.; Liu, X.; Blancato, J.K.; Haddad, B.R.; Wang, W.; Zhong, X.; Choudhary, S.; Krawczyk, E.; Kallakury, B.V.; Davidson, B.J.; et al. Expanding primary cells from mucoepidermoid and other salivary gland neoplasms for genetic and chemosensitivity testing. Dis. Model. Mech. 2018, 11.
- Yuan, H.; Krawczyk, E.; Blancato, J.; Albanese, C.; Zhou, D.; Wang, N.; Paul, S.; Alkhilaiwi, F.; Palechor-Ceron, N.; Dakic, A.; et al. HPV positive neuroendocrine cervical cancer cells are dependent on Myc but not E6/E7 viral oncogenes. Sci. Rep. 2017, 7, 45617.
- Timofeeva, O.A.; Palechor-Ceron, N.; Li, G.; Yuan, H.; Krawczyk, E.; Zhong, X.; Liu, G.; Upadhyay, G.; Dakic, A.; Yu, S.; et al. Conditionally reprogrammed normal and primary tumor prostate epithelial cells: A novel patient-derived cell model for studies of human prostate cancer. Oncotarget 2017, 8, 22741–22758.
- Suprynowicz, F.A.; Kamonjoh, C.M.; Krawczyk, E.; Agarwal, S.; Wellstein, A.; Agboke, F.A.; Choudhury, S.; Liu, X.; Schlegel, R. Conditional cell reprogramming involves non-canonical beta-catenin activation and mTOR-mediated inactivation of Akt. PLoS ONE 2017, 12, e0180897.
- Mahajan, A.S.; Sugita, B.M.; Duttargi, A.N.; Saenz, F.; Krawczyk, E.; McCutcheon, J.N.; Fonseca, A.S.; Kallakury, B.; Pohlmann, P.; Gusev, Y.; et al. Genomic comparison of early-passage conditionally reprogrammed breast cancer cells to their corresponding primary tumors. PLoS ONE 2017, 12, e0186190.
- Chen, C.; Choudhury, S.; Wangsa, D.; Lescott, C.J.; Wilkins, D.J.; Sripadhan, P.; Liu, X.; Wangsa, D.; Ried, T.; Moskaluk, C.; et al. A multiplex preclinical model for adenoid cystic carcinoma of the salivary gland identifies regorafenib as a potential therapeutic drug. Sci. Rep. 2017, 7, 11410.
- Borodovsky, A.; McQuiston, T.J.; Stetson, D.; Ahmed, A.; Whitston, D.; Zhang, J.; Grondine, M.; Lawson, D.; Challberg, S.S.; Zinda, M.; et al. Generation of stable PDX derived cell lines using conditional reprogramming. Mol. Cancer 2017, 16, 177.
