Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Jessie Wu and Version 3 by Jessie Wu.
Predatory bacteria, along with the biology of their predatory behavior, have attracted interest in terms of their ecological significance and industrial applications.
- bacterial predation
- Bdellovibrionota
- BALOs
- predatome
1. Introduction
As comprehensively reviewed by Pérez et al. (2016) [1], predatory bacteria are a group of prokaryotes that can actively hunt and consume other bacteria as their food source. By doing so, they can alter the abundance and diversity of the prey bacteria and thus influence the overall structure of the microbial community. In addition to predatory bacteria, protists and bacteriophages can also have significant impacts on the biomass, structure, and function of microbial communities, though their impacts differ in size, prey specificity, and hunting tactics [2]. Among their interwoven interactions, this mini-researchview, as an update of Pérez et al. (2016) [1], focuses on predatory bacteria with reference to phylogenetic aspects, particularly after the proposal in 2020 of the new phyla Bdellovibrionota and Myxococcota, which show distinct hunting strategies of predation [3].
Pérez et al. (2016) [1] reviewed the hunting strategies of predators of the order Bdellovibrionales, which physically attach to prey cells with flagella-based motility and penetrate into the periplasm of the prey cells, and the order Myxococcales, which are known for a “group attack” with gliding motility, the secretion of lytic enzymes, and the release of antibiotics. Pérez et al. (2016) [1] also reviewed the genomes, transcriptomes, and comparative genomics of predators, including the idea of the “predatome”, i.e., the protein families in phenotypes of predatory bacteria [4]. Through detailed analysis of the predation-related proteins and the encoding genes, predatory properties are predicted for the clades whose predations are not yet known in the phyla Bdellovibrionota and Myxococcota [3]. Moreover, detailed analyses on the correlation between antibiotics biosynthesis and predation indicate that myxobacteria may be prioritized for the discovery of unexplored natural products [5][6][7].
After Pérez et al. (2016) [1], ecological significances and industrial applications of predatory bacteria have been increasingly studied. For example, a study on the potential use of predatory bacteria as alternatives to antibiotics showed that intrarectal inoculations of Bdellovibrio bacteriovorus and Micavibrio aeruginosavorus lead to beneficial and adverse changes, respectively, in rat gut microflora, indicating a top-down control [8]. A large-scale field study using stable isotopes 18O and 13C demonstrated that activities of obligate predators are increased by substrates added to preys, indicating a bottom-up trophic control [9]. A high-resolution microscopic study revealed the submillimeter-scale changes in Vibrio cholerae biofilms attacked by Bdellovibrio bacteriovorus [10]. A recent review evaluates that potential uses of Bdellovibrio and like organisms (BALOs) in medical, agricultural, biotechnological, and environmental applications are achievable and should be pursued [11].
24. Phylogenetic Tree of Predatory Bacteria
A total of 136 sequences of predatory bacterial 16S rRNA genes were collected from 12 phyla (including candidate phyla) of Actinobacteriota, Bacteroidota, Bdellovibrionota, Chloroflexota, Cyanobacteria, Desulfobacteriota, Myxococcota, Ca. Omnitrophica (OP3), Ca. Patescibacteria (CPR) or Ca. Absconditabacteria (SR1), Planctomycetota, Pseudomonadota, and Ca. Saccharibacteria (TM7). The available 16S rRNA gene sequence of “Ca. Vampirococcus lugosii” (accession number MW286273, 1071 bp) [12] was the shortest among the collected sequences, and the phylogenetic trees with and without “Ca. V. lugosii” were constructed, along with the 35 reference sequences from current and former bacterial phyla. The sequences were aligned online with MEGA11 (https://www.megasoftware.net/; accessed on 20 May 2023) [13], and the phylogenetic trees based on the maximum likelihood method were drawn online with iTOL v6 (https://itol.embl.de/; accessed on 20 May 2023) [14] (Figure 1). Figure 1 shows the phylogenetic tree based on the sequences of about 1.6 kb after alignment excluding the shortest 1071 bp sequence of “Ca. V. lugosii”. The 1.6 kb length, instead of the generally cited 1.5 kb, resulted from the alignment of 135 (136 minus 1, Ca. V. lugosii) full-length and near-full-length sequences that contain “gaps”. The tree based on sequences of about 0.6 kb including “Ca. V. lugosii” is diplayed. Information about the used 16S rRNA sequences of predatory bacteria [4][12][15][16][17][18][19][20][21][22][23][24][25][26][27][28][29][30][31][32][33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64][65][66][67][68][69][70][71][72][73][74] are listed along with the hunting strategies of the corresponding predators. The reference 16S rRNA gene sequences from 35 representative, current, and former bacterial phyla [75][76][77][78][79][80][81][82][83][84][85][86][87][88][89][90][91][92][93][94][95][96][97][98][99][100][101][102][103][104][105][106][107][108] are listed. The tree based on “1.6 kb-long” 16S rRNA gene sequences (Figure 1) shows (1) rather nonstreamlined phylogeny and predation strategies in the Bdellovibrionota (orange) and Pseudomonadota (pale pink) cluster from about 3:15 to 5:25 when Figure 1 is seen as the disk display of a 12 h clock and (2) streamlined phylogeny and predation strategies in the Myxococcata cluster (red), except the betaproteobacterial “AF005994 Aristabacter necator” [67] at 10:30. Interestingly, “AF005994 Aristabacter necator” was very deep-branched at about 10 o’clock on the “0.6 kb-long 16S-tree”. The shortest sequence of “MW286273 Vampirococcus lugosii” [12] at about 8:30 on the “0.6 kb long 16S tree”, neighbored with “OM390184 Nanosynbacter lyticus” [17]. Different from Figure 1, The nonpredatory “NR_042149 Fibrobacter succinogenes” [94] clustered with “AB540021 Oligoflexus tunisiensis” [82], which is predicted to be predatory [3] but as yet unconfirmed (Nakai, pers. comm.). These inconsistencies may be a hint for hunting novel predatory bacteria. Another notable irregularity is “CP075895 Chitinophagaceae bacterium” at 9:10 in Figure 1 and 9:05. This bacterium, Ca. Cyanoraptor togatus LGM1 [22], is the only known obligatory predator within the phylum Bacteroidota and is the only known endobiotic invader outside the Bdellovibrionota (orange)–Pseudomonadota (pale pink) cluster from a little before 3:00 to about 5:25. It is also the first obligatory, intracellular predator of cyanobacteria. Only some of the predatory species of the phyla Bdelovibrionota and Myxococcota are shown in Figure 1, which would have exhibited more significant proportions of the phyla if all the predatory species were included. However, the importance of the phyla in the phylogeny of predatory bacteria is already explicit in the current Figure 1 with only selected species.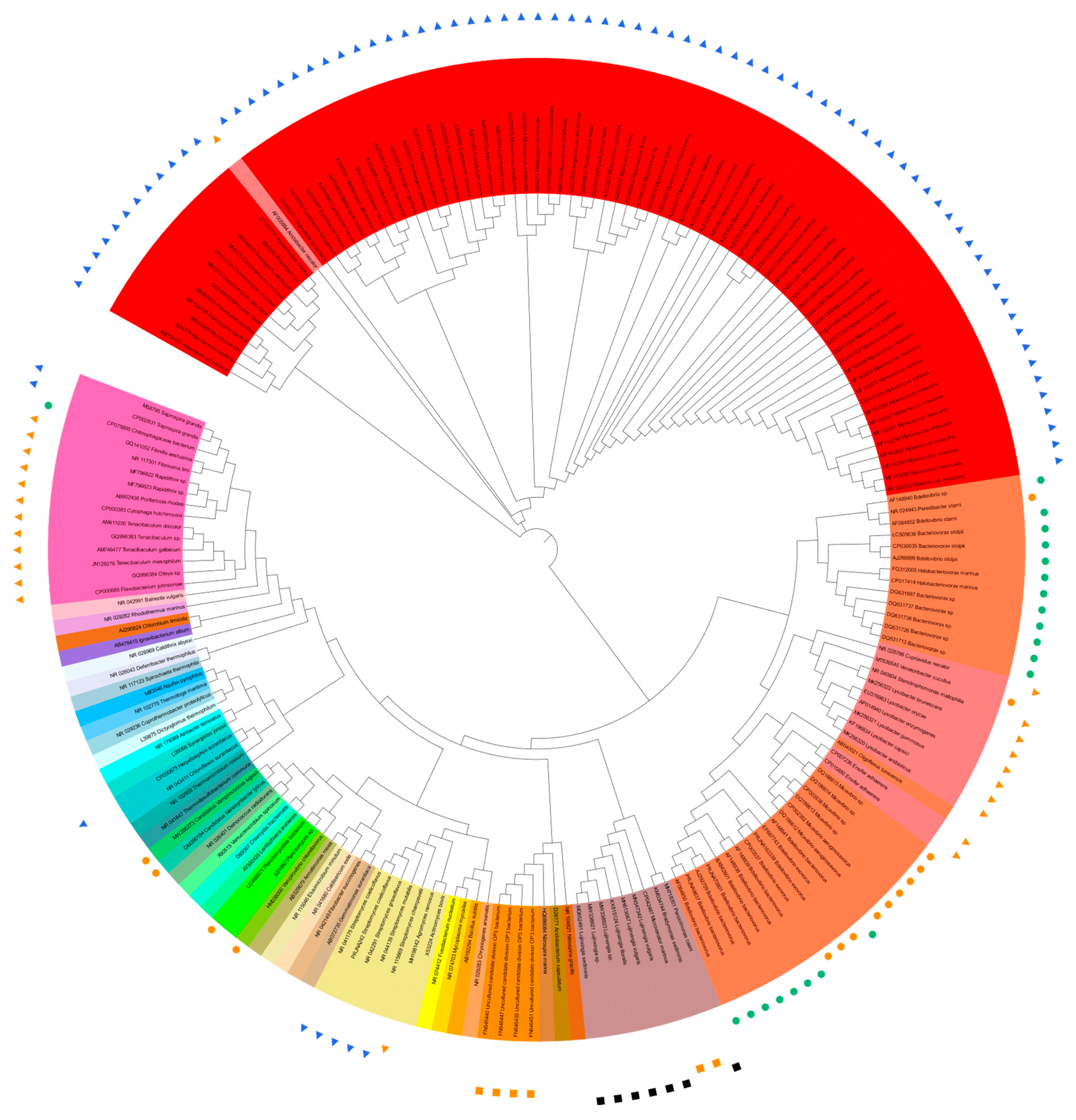

Figure 1. Trans-phylum phylogenetic tree of 135 sequences of predatory bacterial 16S rRNA genes listed [4][12][15][16][17][18][19][20][21][22][23][24][25][26][27][28][29][30][31][32][33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64][65][66][67][68][69][70][71][72][73][74], except [12] and 35 sequences from representative, current, and former bacterial phyla listed [75][76][77][78][79][80][81][82][83][84][85][86][87][88][89][90][91][92][93][94][95][96][97][98][99][100][101][102][103][104][105][106][107][108]. Predation properties are indicated by the symbols as follows: ●, obligate, epibiotic; ●, obligate, endobiotic or direct invasion; ▲, opportunistic, epibiotic; ▲, opportunistic, group attack; ■, facultative, epibiotic; and, ■, facultative, unknown.
References
- Pérez, J.; Moraleda-Munoz, A.; Marcos-Torres, F.J.; Muñoz-Dorado, J. Bacterial predation: 75 years and counting! Environ. Microbiol. 2016, 18, 766–779.
- Johnke, J.; Cohen, Y.; de Leeuw, M.; Kushmaro, A.; Jurkevitch, E.; Chatzinotas, A. Multiple micro-predators controlling bacterial communities in the environment. Curr. Opin. Biotechnol. 2014, 27, 185–190.
- Waite, D.W.; Chuvochina, M.; Pelikan, C.; Parks, D.H.; Yilmaz, P.; Wagner, M.; Loy, A.; Naganuma, T.; Nakai, R.; Whitman, W.B.; et al. Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int. J. Syst. Evol. Microbiol. 2020, 70, 5972–6016.
- Pasternak, Z.; Pietrokovski, S.; Rotem, O.; Gophna, U.; Lurie-Weinberger, M.N.; Jurkevitch, E. By their genes ye shall know them: Genomic signatures of predatory bacteria. ISME J. 2012, 7, 756–769.
- Korp, J.; Gurovic, M.S.V.; Nett, M. Antibiotics from predatory bacteria. Beilstein J. Org. Chem. 2016, 12, 594–607.
- Gregory, K.; Salvador, L.A.; Akbar, S.; Adaikpoh, B.I.; Stevens, D.C. Survey of Biosynthetic Gene Clusters from Sequenced Myxobacteria Reveals Unexplored Biosynthetic Potential. Microorganisms 2019, 7, 181.
- Ahearne, A.; Albataineh, H.; Dowd, S.E.; Stevens, D.C. Assessment of Evolutionary Relationships for Prioritization of Myxobacteria for Natural Product Discovery. Microorganisms 2021, 9, 1376.
- Shatzkes, K.; Tang, C.; Singleton, E.; Shukla, S.; Zuena, M.; Gupta, S.; Dharani, S.; Rinaggio, J.; Connell, N.D.; Kadouri, D.E. Effect of predatory bacteria on the gut bacterial microbiota in rats. Sci. Rep. 2017, 7, srep43483.
- Hungate, B.A.; Marks, J.C.; Power, M.E.; Schwartz, E.; van Groenigen, K.J.; Blazewicz, S.J.; Chuckran, P.; Dijkstra, P.; Finley, B.K.; Firestone, M.K.; et al. The Functional Significance of Bacterial Predators. mBio 2021, 12, e00466-21.
- Wucher, B.R.; Elsayed, M.; Adelman, J.S.; Kadouri, D.E.; Nadell, C.D. Bacterial predation transforms the landscape and community assembly of biofilms. Curr. Biol. 2021, 31, 2643–2651.e3.
- Mookherjee, A.; Jurkevitch, E. Interactions between Bdellovibrio and like organisms and bacteria in biofilms: Beyond predator–prey dynamics. Environ. Microbiol. 2021, 24, 998–1011.
- Moreira, D.; Zivanovic, Y.; López-Archilla, A.I.; Iniesto, M.; López-García, P. Reductive evolution and unique predatory mode in the CPR bacterium Vampirococcus lugosii. Nat. Commun. 2021, 12, 1–11.
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027.
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296.
- Li, N.; Wang, K.; Williams, H.N.; Sun, J.; Ding, C.; Leng, X.; Dong, K. Analysis of gene gain and loss in the evolution of predatory bacteria. Gene 2017, 598, 63–70.
- Enos, B.G.; Anthony, M.K.; DeGiorgis, J.A.; Williams, L.E. Prey Range and Genome Evolution of Halobacteriovorax marinus Predatory Bacteria from an Estuary. Msphere 2018, 3, e00508-17.
- He, X.; McLean, J.S.; Edlund, A.; Yooseph, S.; Hall, A.P.; Liu, S.-Y.; Dorrestein, P.C.; Esquenazi, E.; Hunter, R.C.; Cheng, G.; et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc. Natl. Acad. Sci. USA 2014, 112, 244–249.
- Inoue, D.; Hiroshima, N.; Nakamura, S.; Ishizawa, H.; Ike, M. Characterization of Two Novel Predatory Bacteria, Bacteriovorax stolpii HI3 and Myxococcus sp. MH1, Isolated from a Freshwater Pond: Prey Range, and Predatory Dynamics and Efficiency. Microorganisms 2022, 10, 1816.
- Shiratori, T.; Suzuki, S.; Kakizawa, Y.; Ishida, K.-I. Phagocytosis-like cell engulfment by a planctomycete bacterium. Nat. Commun. 2019, 10, 5529.
- Wang, S.; Mu, D.; Du, Z.-J. Persicimonas caeni gen. nov., sp. nov., the Representative of a Novel Wide-Ranging Predatory Taxon in Bradymonadales. Front. Microbiol. 2020, 11, 698.
- Guo, L.-Y.; Li, C.-M.; Wang, S.; Mu, D.-S.; Du, Z.-J. Lujinxingia litoralis gen. nov., sp. nov. and Lujinxingia sediminis sp. nov., two new representatives in the order Bradymonadales. Int. J. Syst. Evol. Microbiol. 2019, 69, 2767–2774.
- Bethany, J.; Johnson, S.L.; Garcia-Pichel, F. High impact of bacterial predation on cyanobacteria in soil biocrusts. Nat. Commun. 2022, 13, 1–10.
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.-M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147.
- Ibrahimi, M.; Korichi, W.; Hafidi, M.; Lemee, L.; Ouhdouch, Y.; Loqman, S. Marine Actinobacteria: Screening for Predation Leads to the Discovery of Potential New Drugs against Multidrug-Resistant Bacteria. Antibiotics 2020, 9, 91.
- Mahler, L.; Wink, K.; Beulig, R.J.; Scherlach, K.; Tovar, M.; Zang, E.; Martin, K.; Hertweck, C.; Belder, D.; Roth, M. Detection of antibiotics synthetized in microfluidic picolitre-droplets by various actinobacteria. Sci. Rep. 2018, 8, 13087.
- Schwudke, D.; Strauch, E.; Krueger, M.; Appel, B. Taxonomic Studies of Predatory Bdellovibrios Based on 16S rRNA Analysis, Ribotyping and the hit Locus and Characterization of Isolates from the Gut of Animals. Syst. Appl. Microbiol. 2001, 24, 385–394.
- Crossman, L.C.; Chen, H.; Cerdeño-Tárraga, A.-M.; Brooks, K.; Quail, M.A.; Pineiro, S.A.; Hobley, L.; Sockett, R.E.; Bentley, S.D.; Parkhill, J.; et al. A small predatory core genome in the divergent marine Bacteriovorax marinus SJ and the terrestrial Bdellovibrio bacteriovorus. ISME J. 2012, 7, 148–160.
- Pineiro, S.A.; Stine, O.C.; Chauhan, A.; Steyert, S.R.; Smith, R.; Williams, H.N. Global survey of diversity among environmental saltwater Bacteriovoracaceae. Environ. Microbiol. 2007, 9, 2441–2450.
- Baer, M.L.; Ravel, J.; Chun, J.; Hill, R.T.; Williams, H.N. A proposal for the reclassification of Bdellovibrio stolpii and Bdellovibrio starrii into a new genus, Bacteriovorax gen. nov. as Bacteriovorax stolpii comb. nov. and Bacteriovorax starrii comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 2000, 50, 219–224.
- Jurkevitch, E.; Minz, D.; Ramati, B.; Barel, G. Prey Range Characterization, Ribotyping, and Diversity of Soil and Rhizosphere Bdellovibrio spp. Isolated on Phytopathogenic Bacteria. Appl. Environ. Microbiol. 2000, 66, 2365–2371.
- Rendulic, S.; Jagtap, P.; Rosinus, A.; Eppinger, M.; Baar, C.; Lanz, C.; Keller, H.; Lambert, C.; Evans, K.J.; Goesmann, A.; et al. A Predator Unmasked: Life Cycle of Bdellovibrio bacteriovorus from a Genomic Perspective. Science 2004, 303, 689–692.
- Koval, S.F.; Hynes, S.H.; Flannagan, R.S.; Pasternak, Z.; Davidov, Y.; Jurkevitch, E. Bdellovibrio exovorus sp. nov., a novel predator of Caulobacter crescentus. Int. J. Syst. Evol. Microbiol. 2013, 63, 146–151.
- Davidov, Y.; Huchon, D.; Koval, S.F.; Jurkevitch, E. A new ?-proteobacterial clade of Bdellovibrio-like predators: Implications for the mitochondrial endosymbiotic theory. Environ. Microbiol. 2006, 8, 2179–2188.
- Wang, Z.; Kadouri, D.E.; Wu, M. Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genom. 2011, 12, 453.
- Gherna, R.; Woese, C. A Partial Phylogenetic Analysis of the “Flavobacter-Bacteroides” Phylum: Basis for Taxonomic Restructuring. Syst. Appl. Microbiol. 1992, 15, 513–521.
- Meincke, L.; Copeland, A.; Lapidus, A.; Lucas, S.; Berry, K.W.; Del Rio, T.G.; Hammon, N.; Dalin, E.; Tice, H.; Pitluck, S.; et al. Complete genome sequence of Polynucleobacter necessarius subsp. asymbioticus type strain (QLW-P1DMWA-1T). Stand. Genom. Sci. 2012, 6, 1–14.
- Xie, G.; Bruce, D.C.; Challacombe, J.F.; Chertkov, O.; Detter, J.C.; Gilna, P.; Han, C.S.; Lucas, S.; Misra, M.; Myers, G.L.; et al. Genome Sequence of the Cellulolytic Gliding Bacterium Cytophaga hutchinsonii. Appl. Environ. Microbiol. 2007, 73, 3536–3546.
- Yoon, J.; Oku, N.; Park, S.; Kasai, H.; Yokota, A. Porifericola rhodea gen. nov., sp. nov., a new member of the phylum Bacteroidetes isolated by the bait-streaked agar technique. Antonie van Leeuwenhoek 2011, 100, 145–153.
- Filippini, M.; Svercel, M.; Laczko, E.; Kaech, A.; Ziegler, U.; Bagheri, H.C. Fibrella aestuarina gen. nov., sp. nov., a filamentous bacterium of the family Cytophagaceae isolated from a tidal flat, and emended description of the genus Rudanella Weon et al. 2008. Int. J. Syst. Evol. Microbiol. 2011, 61, 184–189.
- McBride, M.J.; Xie, G.; Martens, E.C.; Lapidus, A.; Henrissat, B.; Rhodes, R.G.; Goltsman, E.; Wang, W.; Xu, J.; Hunnicutt, D.W.; et al. Novel Features of the Polysaccharide-Digesting Gliding Bacterium Flavobacterium johnsoniae as Revealed by Genome Sequence Analysis. Appl. Environ. Microbiol. 2009, 75, 6864–6875.
- Banning, E.C.; Casciotti, K.L.; Kujawinski, E.B. Novel strains isolated from a coastal aquifer suggest a predatory role for flavobacteria. FEMS Microbiol. Ecol. 2010, 73, 254–270.
- Piñeiro-Vidal, M.; Riaza, A.; Santos, Y. Tenacibaculum discolor sp. nov. and Tenacibaculum gallaicum sp. nov., isolated from sole (Solea senegalensis) and turbot (Psetta maxima) culture systems. Int. J. Syst. Evol. Microbiol. 2008, 58, 21–25.
- Suzuki, M.; Nakagawa, Y.; Harayama, S.; Yamamoto, S. Phylogenetic analysis and taxonomic study of marine Cytophaga-like bacteria: Proposal for Tenacibaculum gen. nov. with Tenacibaculum maritimum comb. nov. and Tenacibaculum ovolyticum comb. nov., and description of Tenacibaculum mesophilum sp. nov. and Tenacibaculum amylolyticum sp. nov. Int. J. Syst. Evol. Microbiol. 2001, 51, 1639–1652.
- Kiss, H.; Nett, M.; Domin, N.; Martin, K.; Maresca, J.A.; Copeland, A.; Lapidus, A.; Lucas, S.; Berry, K.W.; Del Rio, T.G.; et al. Complete genome sequence of the filamentous gliding predatory bacterium Herpetosiphon aurantiacus type strain (114-95T). Stand. Genom. Sci. 2011, 5, 356–370.
- Wang, Z.-J.; Liu, Q.-Q.; Zhao, L.-H.; Du, Z.-J.; Chen, G.-J. Bradymonas sediminis gen. nov., sp. nov., isolated from coastal sediment, and description of Bradymonadaceae fam. nov. and Bradymonadales ord. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 1542–1549.
- Gong, Y.; Ping, X.-Y.; Zeng, C.-H.; Wang, S.-X.; Zhou, Y.; Wang, M.-Y.; Mu, D.-S.; Du, Z.-J. Predation capacity of Bradymonabacteria, a recently discovered group in the order Bradymonadales, isolated from marine sediments. Arch. Microbiol. 2022, 204, 1–11.
- Spröer, C.; Reichenbach, H.; Stackebrandt, E. The correlation between morphological and phylogenetic classification of myxobacteria. Int. J. Syst. Evol. Microbiol. 1999, 49, 1255–1262.
- Shimkets, L.; Woese, C.R. A phylogenetic analysis of the myxobacteria: Basis for their classification. Proc. Natl. Acad. Sci. USA 1992, 89, 9459–9463.
- Huntley, S.; Hamann, N.; Wegener-Feldbrugge, S.; Treuner-Lange, A.; Kube, M.; Reinhardt, R.; Klages, S.; Muller, R.; Ronning, C.M.; Nierman, W.C.; et al. Comparative Genomic Analysis of Fruiting Body Formation in Myxococcales. Mol. Biol. Evol. 2010, 28, 1083–1097.
- Iizuka, T.; Jojima, Y.; Fudou, R.; Yamanaka, S. Isolation of myxobacteria from the marine environment. FEMS Microbiol. Lett. 1998, 169, 317–322.
- Huntley, S.; Zhang, Y.; Treuner-Lange, A.; Kneip, S.; Sensen, C.W.; Søgaard-Andersen, L. Complete Genome Sequence of the Fruiting Myxobacterium Corallococcus coralloides DSM 2259. J. Bacteriol. 2012, 194, 3012–3013.
- Livingstone, P.; Morphew, R.; Whitworth, D.E. Myxobacteria Are Able to Prey Broadly upon Clinically-Relevant Pathogens, Exhibiting a Prey Range Which Cannot Be Explained by Phylogeny. Front. Microbiol. 2017, 8, 1593.
- Schieferdecker, S.; Exner, T.E.; Gross, H.; Roth, M.; Nett, M. New myxothiazols from the predatory bacterium Myxococcus fulvus. J. Antibiot. 2014, 67, 519–525.
- Goldman, B.S.; Nierman, W.C.; Kaiser, D.; Slater, S.C.; Durkin, A.S.; Eisen, J.A.; Ronning, C.M.; Barbazuk, W.B.; Blanchard, M.; Field, C.; et al. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. USA 2006, 103, 15200–15205.
- Jain, R.; Habermann, B.H.; Mignot, T. Complete Genome Assembly of Myxococcus xanthus Strain DZ2 Using Long High-Fidelity (HiFi) Reads Generated with PacBio Technology. Genome Announc. 2021, 10, e0053021.
- Oyaizu, H.; Woese, C. Phylogenetic Relationships Among the Sulfate Respiring Bacteria, Myxobacteria and Purple Bacteria. Syst. Appl. Microbiol. 1985, 6, 257–263.
- Vos, M.; Velicer, G.J. Genetic Population Structure of the Soil Bacterium Myxococcus xanthus at the Centimeter Scale. Appl. Environ. Microbiol. 2006, 72, 3615–3625.
- Garcia, R.; Gerth, K.; Stadler, M.; Dogma, I.J.; Müller, R. Expanded phylogeny of myxobacteria and evidence for cultivation of the ‘unculturables’. Mol. Phylogenetics Evol. 2010, 57, 878–887.
- Iizuka, T.; Jojima, Y.; Fudou, R.; Hiraishi, A.; Ahn, J.-W.; Yamanaka, S. Plesiocystis pacifica gen. nov., sp. nov., a marine myxobacterium that contains dihydrogenated menaquinone, isolated from the Pacific coasts of Japan. Int. J. Syst. Evol. Microbiol. 2003, 53, 189–195.
- Zaburannyi, N.; Bunk, B.; Maier, J.; Overmann, J.; Müller, R. Genome Analysis of the Fruiting Body-Forming Myxobacterium Chondromyces crocatus Reveals High Potential for Natural Product Biosynthesis. Appl. Environ. Microbiol. 2016, 82, 1945–1957.
- Garcia, R.; Gemperlein, K.; Müller, R. Minicystis rosea gen. nov., sp. nov., a polyunsaturated fatty acid-rich and steroid-producing soil myxobacterium. Int. J. Syst. Evol. Microbiol. 2014, 64, 3733–3742.
- Awal, R.P.; Garcia, R.; Müller, R. Racemicystis crocea gen. nov., sp. nov., a soil myxobacterium in the family Polyangiaceae. Int. J. Syst. Evol. Microbiol. 2016, 66, 2389–2395.
- Schneiker, S.; Perlova, O.; Kaiser, O.; Gerth, K.; Alici, A.; Altmeyer, M.O.; Bartels, D.; Bekel, T.; Beyer, S.; Bode, E.; et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat. Biotechnol. 2007, 25, 1281–1289.
- Rotaru, A.-E. Visualization of Candidate Division OP3 Cocci in Limonene-Degrading Methanogenic Cultures. J. Microbiol. Biotechnol. 2012, 22, 457–461.
- Williams, L.E.; Baltrus, D.A.; O’donnell, S.D.; Skelly, T.J.; Martin, M.O. Complete Genome Sequence of the Predatory Bacterium Ensifer adhaerens Casida A. Genome Announc. 2017, 5, e01344-17.
- Rudder, S.; Doohan, F.; Creevey, C.J.; Wendt, T.; Mullins, E. Genome sequence of Ensifer adhaerens OV14 provides insights into its ability as a novel vector for the genetic transformation of plant genomes. BMC Genom. 2014, 15, 268.
- Cain, C.C.; Lee, D.; Waldo, R.H.; Henry, A.T.; Casida, E.J.; Wani, M.C.; Wall, M.E.; Oberlies, N.H.; Falkinham, J.O. Synergistic Antimicrobial Activity of Metabolites Produced by a Nonobligate Bacterial Predator. Antimicrob. Agents Chemother. 2003, 47, 2113–2117.
- Poehlein, A.; Kusian, B.; Friedrich, B.; Daniel, R.; Bowien, B. Complete Genome Sequence of the Type Strain Cupriavidus necator N-1. J. Bacteriol. 2011, 193, 5017.
- Saeedi, A.; Cummings, N.J.; McLean, D.; Connerton, I.F.; Connerton, P.L. Venatorbacter cucullus gen. nov sp. nov a novel bacterial predator. Sci. Rep. 2021, 11, 21393.
- Zhao, Y.; Jiang, T.; Xu, H.; Xu, G.; Qian, G.; Liu, F. Characterization of Lysobacter spp. strains and their potential use as biocontrol agents against pear anthracnose. Microbiol. Res. 2020, 242, 126624.
- Puopolo, G.; Giovannini, O.; Pertot, I. Lysobacter capsici AZ78 can be combined with copper to effectively control Plasmopara viticola on grapevine. Microbiol. Res. 2014, 169, 633–642.
- Takami, H.; Toyoda, A.; Uchiyama, I.; Itoh, T.; Takaki, Y.; Arai, W.; Nishi, S.; Kawai, M.; Shin-Ya, K.; Ikeda, H. Complete genome sequence and expression profile of the commercial lytic enzyme producer Lysobacter enzymogenes M497-1. DNA Res. 2017, 24, 169–177.
- Aslam, Z.; Yasir, M.; Jeon, C.O.; Chung, Y.R. Lysobacter oryzae sp. nov., isolated from the rhizosphere of rice (Oryza sativa L.). Int. J. Syst. Evol. Microbiol. 2009, 59, 675–680.
- Anzai, Y.; Kim, H.; Park, J.-Y.; Wakabayashi, H.; Oyaizu, H. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 2000, 50, 1563–1589.
- Hiraishi, A.; Kishimoto, N.; Kosako, Y.; Wakao, N.; Tano, T. Phylogenetic position of the menaquinone-containing acidophilic chemo-organotrophAcidobacterium capsulatum. FEMS Microbiol. Lett. 1995, 132, 91–94.
- Stackebrandt, E.; Charfreitag, O. Partial 16S rRNA primary structure of five Actinomyces species: Phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J. Gen. Microbiol. 1990, 136, 37–43.
- Burggraf, S.; Olsen, G.; Stetter, K.; Woese, C. A Phylogenetic Analysis of Aquifex pyrophilus. Syst. Appl. Microbiol. 1992, 15, 352–356.
- Tamaki, H.; Tanaka, Y.; Matsuzawa, H.; Muramatsu, M.; Meng, X.-Y.; Hanada, S.; Mori, K.; Kamagata, Y. Armatimonas rosea gen. nov., sp. nov., of a novel bacterial phylum, Armatimonadetes phyl. nov., formally called the candidate phylum OP10. Int. J. Syst. Evol. Microbiol. 2011, 61, 1442–1447.
- Katayama, T.; Nobu, M.K.; Kusada, H.; Meng, X.-Y.; Hosogi, N.; Uematsu, K.; Yoshioka, H.; Kamagata, Y.; Tamaki, H. Isolation of a member of the candidate phylum ‘Atribacteria’ reveals a unique cell membrane structure. Nat. Commun. 2020, 11, 6381.
- Setyorini, E.; Kim, Y.-J.; Takenaka, S.; Murakami, S.; Aoki, K. Purification and characterization of a halotolerant intracellular protease fromBacillus subtilis strain FP-133. J. Basic Microbiol. 2006, 46, 294–304.
- Urios, L.; Agogué, H.; Lesongeur, F.; Stackebrandt, E.; Lebaron, P. Balneola vulgaris gen. nov., sp. nov., a member of the phylum Bacteroidetes from the north-western Mediterranean Sea. Int. J. Syst. Evol. Microbiol. 2006, 56, 1883–1887.
- Nakai, R.; Nishijima, M.; Tazato, N.; Handa, Y.; Karray, F.; Sayadi, S.; Isoda, H.; Naganuma, T. Oligoflexus tunisiensis gen. nov., sp. nov., a Gram-negative, aerobic, filamentous bacterium of a novel proteobacterial lineage, and description of Oligoflexaceae fam. nov., Oligoflexales ord. nov. and Oligoflexia classis nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 3353–3359.
- Mori, K.; Yamaguchi, K.; Sakiyama, Y.; Urabe, T.; Suzuki, K.-I. Caldisericum exile gen. nov., sp. nov., an anaerobic, thermophilic, filamentous bacterium of a novel bacterial phylum, Caldiserica phyl. nov., originally called the candidate phylum OP5, and description of Caldisericaceae fam. nov., Caldisericales ord. nov. and Caldisericia classis nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 2894–2898.
- Miroshnichenko, M.L.; Kostrikina, N.A.; Chernyh, N.A.; Pimenov, N.V.; Tourova, T.P.; Antipov, A.N.; Spring, S.; Stackebrandt, E.; Bonch-Osmolovskaya, E.A. Caldithrix abyssi gen. nov., sp. nov., a nitrate-reducing, thermophilic, anaerobic bacterium isolated from a Mid-Atlantic Ridge hydrothermal vent, represents a novel bacterial lineage. Int. J. Syst. Evol. Microbiol. 2003, 53, 323–329.
- Tmoko, P.; Fukushi, H.; Ochiai, Y.; Yamaguchi, T.; Hirai, K. Phylogenetic Analysis of the Genus Chlamydia Based on 16S rRNA Gene Sequences. Int. J. Syst. Evol. Microbiol. 1997, 47, 425–431.
- Alexander, B.; Andersen, J.H.; Cox, R.P.; Imhoff, J.F. Phylogeny of green sulfur bacteria on the basis of gene sequences of 16S rRNA and of the Fenna-Matthews-Olson protein. Arch. Microbiol. 2002, 178, 131–140.
- Hanada, S.; Hiraishi, A.; Shimada, K.; Matsuura, K. Chloroflexus aggregans sp. nov., a Filamentous Phototrophic Bacterium Which Forms Dense Cell Aggregates by Active Gliding Movement. Int. J. Syst. Evol. Microbiol. 1995, 45, 676–681.
- Macy, J.M.; Nunan, K.; Hagen, K.D.; Dixon, D.R.; Harbour, P.J.; Cahill, M.; Sly, L.I. Chrysiogenes arsenatis gen. nov., sp. nov., a New Arsenate-Respiring Bacterium Isolated from Gold Mine Wastewater. Int. J. Syst. Evol. Microbiol. 1996, 46, 1153–1157.
- Alexiev, A.; Coil, D.A.; Badger, J.H.; Enticknap, J.; Ward, N.; Robb, F.T.; Eisen, J.A. Complete Genome Sequence of Coprothermobacter proteolyticus DSM 5265. Genome Announc. 2014, 2, e00470-14.
- Greene, A.C.; Patel, B.K.C.; Sheehy, A.J. Deferribacter thermophilus gen. nov., sp. nov., a Novel Thermophilic Manganese- and Iron-Reducing Bacterium Isolated from a Petroleum Reservoir. Int. J. Syst. Evol. Microbiol. 1997, 47, 505–509.
- Rainey, F.A.; Nobre, M.F.; Schumann, P.; Stackebrandt, E.; DA Costa, M.S. Phylogenetic Diversity of the Deinococci as Determined by 16S Ribosomal DNA Sequence Comparison. Int. J. Syst. Evol. Microbiol. 1997, 47, 510–514.
- Gibbs, M.D.; Reeves, R.A.; Bergquist, P.L. Cloning, sequencing, and expression of a xylanase gene from the extreme thermophile Dictyoglomus thermophilum Rt46B.1 and activity of the enzyme on fiber-bound substrate. Appl. Environ. Microbiol. 1995, 61, 4403–4408.
- Geissinger, O.; Herlemann, D.P.R.; Moörschel, E.; Maier, U.G.; Brune, A. The Ultramicrobacterium “Elusimicrobium minutum” gen. nov., sp. nov., the First Cultivated Representative of the Termite Group 1 Phylum. Appl. Environ. Microbiol. 2009, 75, 2831–2840.
- Beéra-Maillet, C.; Ribot, Y.; Forano, E. Fiber-Degrading Systems of Different Strains of the Genus Fibrobacter. Appl. Environ. Microbiol. 2004, 70, 2172–2179.
- Kapatral, V.; Anderson, I.; Ivanova, N.; Reznik, G.; Los, T.; Lykidis, A.; Bhattacharyya, A.; Bartman, A.; Gardner, W.; Grechkin, G.; et al. Genome Sequence and Analysis of the Oral Bacterium Fusobacterium nucleatum Strain ATCC 25586. J. Bacteriol. 2002, 184, 2005–2018.
- Zhang, H.; Sekiguchi, Y.; Hanada, S.; Hugenholtz, P.; Kim, H.; Kamagata, Y.; Nakamura, K. Gemmatimonas aurantiaca gen. nov., sp. nov., a Gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 1155–1163.
- Iino, T.; Mori, K.; Uchino, Y.; Nakagawa, T.; Harayama, S.; Suzuki, K.-I. Ignavibacterium album gen. nov., sp. nov., a moderately thermophilic anaerobic bacterium isolated from microbial mats at a terrestrial hot spring and proposal of Ignavibacteria classis nov., for a novel lineage at the periphery of green sulfur bacteria. Int. J. Syst. Evol. Microbiol. 2010, 60, 1376–1382.
- Cho, J.-C.; Vergin, K.L.; Morris, R.M.; Giovannoni, S.J. Lentisphaera araneosa gen. nov., sp. nov, a transparent exopolymer producing marine bacterium, and the description of a novel bacterial phylum, Lentisphaerae. Environ. Microbiol. 2004, 6, 611–621.
- Westberg, J.; Persson, A.; Holmberg, A.; Goesmann, A.; Lundeberg, J.; Johansson, K.-E.; Pettersson, B.; Uhlén, M. The Genome Sequence of Mycoplasma mycoides subsp. mycoides SC Type Strain PG1T, the Causative Agent of Contagious Bovine Pleuropneumonia (CBPP). Genome Res. 2004, 14, 221–227.
- Yarza, P.; Spröer, C.; Swiderski, J.; Mrotzek, N.; Spring, S.; Tindall, B.J.; Gronow, S.; Pukall, R.; Klenk, H.-P.; Lang, E.; et al. Sequencing orphan species initiative (SOS): Filling the gaps in the 16S rRNA gene sequence database for all species with validly published names. Syst. Appl. Microbiol. 2013, 36, 69–73.
- Keuter, S.; Kruse, M.; Lipski, A.; Spieck, E. Relevance of Nitrospira for nitrite oxidation in a marine recirculation aquaculture system and physiological features of a Nitrospira marina-like isolate. Environ. Microbiol. 2011, 13, 2536–2547.
- Ward, N.; Rainey, F.A.; Stackebrandt, E.; Schlesner, H. Unraveling the extent of diversity within the order Planctomycetales. Appl. Environ. Microbiol. 1995, 61, 2270–2275.
- Andrésson, O.S.; Fridjónsson, O.H. The sequence of the single 16S rRNA gene of the thermophilic eubacterium Rhodothermus marinus reveals a distant relationship to the group containing Flexibacter, Bacteroides, and Cytophaga species. J. Bacteriol. 1994, 176, 6165–6169.
- Allison, M.J.; Mayberry, W.R.; Mcsweeney, C.S.; Stahl, D.A. Synergistes jonesii, gen. nov., sp.nov.: A Rumen Bacterium That Degrades Toxic Pyridinediols. Syst. Appl. Microbiol. 1992, 15, 522–529.
- Friedrich, M.W. Phylogenetic Analysis Reveals Multiple Lateral Transfers of Adenosine-5′-Phosphosulfate Reductase Genes among Sulfate-Reducing Microorganisms. J. Bacteriol. 2002, 184, 278–289.
- Wu, D.; Raymond, J.; Wu, M.; Chatterji, S.; Ren, Q.; Graham, J.E.; Bryant, D.A.; Robb, F.; Colman, A.; Tallon, L.J.; et al. Complete Genome Sequence of the Aerobic CO-Oxidizing Thermophile Thermomicrobium roseum. PLoS ONE 2009, 4, e4207.
- Nelson, K.E.; Clayton, R.A.; Gill, S.R.; Gwinn, M.L.; Dodson, R.J.; Haft, D.H.; Hickey, E.K.; Peterson, J.D.; Nelson, W.C.; Ketchum, K.A.; et al. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 1999, 399, 323–329.
- Ward-Rainey, N.; Rainey, F.A.; Schlesner, H.; Stackebrandt, E. Assignment of hitherto unidentified 16S rDNA species to a main line of descent within the domain Bacteria. Microbiology 1995, 141, 3247–3250.
- Phillips, K.E.; Akbar, S.; Stevens, D.C. Concepts and conjectures concerning predatory performance of myxobacteria. Front. Microbiol. 2022, 13.
More
