Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Takeshi Naganuma and Version 3 by Jessie Wu.
Predatory bacteria, along with the biology of their predatory behavior, have attracted interest in terms of their ecological significance and industrial applications.
- bacterial predation
- Bdellovibrionota
- BALOs
- predatome
1. Introduction
As comprehensively reviewed by Pérez et al. (2016) [1], predatory bacteria are a group of prokaryotes that can actively hunt and consume other bacteria as their food source. By doing so, they can alter the abundance and diversity of the prey bacteria and thus influence the overall structure of the microbial community. In addition to predatory bacteria, protists and bacteriophages can also have significant impacts on the biomass, structure, and function of microbial communities, though their impacts differ in size, prey specificity, and hunting tactics [2]. Among their interwoven interactions, this mini-researchview, as an update of Pérez et al. (2016) [1], focuses on predatory bacteria with reference to phylogenetic aspects, particularly after the proposal in 2020 of the new phyla Bdellovibrionota and Myxococcota, which show distinct hunting strategies of predation [3].
Pérez et al. (2016) [1] reviewed the hunting strategies of predators of the order Bdellovibrionales, which physically attach to prey cells with flagella-based motility and penetrate into the periplasm of the prey cells, and the order Myxococcales, which are known for a “group attack” with gliding motility, the secretion of lytic enzymes, and the release of antibiotics. Pérez et al. (2016) [1] also reviewed the genomes, transcriptomes, and comparative genomics of predators, including the idea of the “predatome”, i.e., the protein families in phenotypes of predatory bacteria [4]. Through detailed analysis of the predation-related proteins and the encoding genes, predatory properties are predicted for the clades whose predations are not yet known in the phyla Bdellovibrionota and Myxococcota [3]. Moreover, detailed analyses on the correlation between antibiotics biosynthesis and predation indicate that myxobacteria may be prioritized for the discovery of unexplored natural products [5][6][7][5,6,7].
After Pérez et al. (2016) [1], ecological significances and industrial applications of predatory bacteria have been increasingly studied. For example, a study on the potential use of predatory bacteria as alternatives to antibiotics showed that intrarectal inoculations of Bdellovibrio bacteriovorus and Micavibrio aeruginosavorus lead to beneficial and adverse changes, respectively, in rat gut microflora, indicating a top-down control [8]. A large-scale field study using stable isotopes 18O and 13C demonstrated that activities of obligate predators are increased by substrates added to preys, indicating a bottom-up trophic control [9]. A high-resolution microscopic study revealed the submillimeter-scale changes in Vibrio cholerae biofilms attacked by Bdellovibrio bacteriovorus [10]. A recent review evaluates that potential uses of Bdellovibrio and like organisms (BALOs) in medical, agricultural, biotechnological, and environmental applications are achievable and should be pursued [11].
24. Phylogenetic Tree of Predatory Bacteria
A total of 136 sequences of predatory bacterial 16S rRNA genes were collected from 12 phyla (including candidate phyla) of Actinobacteriota, Bacteroidota, Bdellovibrionota, Chloroflexota, Cyanobacteria, Desulfobacteriota, Myxococcota, Ca. Omnitrophica (OP3), Ca. Patescibacteria (CPR) or Ca. Absconditabacteria (SR1), Planctomycetota, Pseudomonadota, and Ca. Saccharibacteria (TM7). The available 16S rRNA gene sequence of “Ca. Vampirococcus lugosii” (accession number MW286273, 1071 bp) [12][37] was the shortest among the collected sequences, and the phylogenetic trees with and without “Ca. V. lugosii” were constructed, along with the 35 reference sequences from current and former bacterial phyla. The sequences were aligned online with MEGA11 (https://www.megasoftware.net/; accessed on 20 May 2023) [13][67], and the phylogenetic trees based on the maximum likelihood method were drawn online with iTOL v6 (https://itol.embl.de/; accessed on 20 May 2023) [14][68] (Figure 1 and Figure S1). Figure 1 shows the phylogenetic tree based on the sequences of about 1.6 kb after alignment excluding the shortest 1071 bp sequence of “Ca. V. lugosii”. The 1.6 kb length, instead of the generally cited 1.5 kb, resulted from the alignment of 135 (136 minus 1, Ca. V. lugosii) full-length and near-full-length sequences that contain “gaps”. TFigure S1 displays the tree based on sequences of about 0.6 kb including “Ca. V. lugosii” is diplayed. Information about the used 16S rRNA sequences of predatory bacteria [4][12][15][16][17][18][19][20][21][22][23][24][25][26][27][28][29][30][31][32][33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64][65][66][67][68][69][70][71][72][73][74][4,18,20,31,37,42,47,49,50,61,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120] are listed in Table S1 along with the hunting strategies of the corresponding predators. The reference 16S rRNA gene sequences from 35 representative, current, and former bacterial phyla [75][76][77][78][79][80][81][82][83][84][85][86][87][88][89][90][91][92][93][94][95][96][97][98][99][100][101][102][103][104][105][106][107][108][121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154] are listed in Table S2. The tree based on “1.6 kb-long” 16S rRNA gene sequences (Figure 1) shows (1) rather nonstreamlined phylogeny and predation strategies in the Bdellovibrionota (orange) and Pseudomonadota (pale pink) cluster from about 3:15 to 5:25 when Figure 1 is seen as the disk display of a 12 h clock and (2) streamlined phylogeny and predation strategies in the Myxococcata cluster (red), except the betaproteobacterial “AF005994 Aristabacter necator” [67][113] at 10:30. Interestingly, “AF005994 Aristabacter necator” was very deep-branched at about 10 o’clock on the “0.6 kb-long 16S-tree”, as shown in Figure S1. TFigure S1 (0.6 kb long 16S tree) includes the shortest sequence of “MW286273 Vampirococcus lugosii” [12][37] at about 8:30 on the “0.6 kb long 16S tree”, neighbored with “OM390184 Nanosynbacter lyticus” [17][31]. Different from Figure 1, TFigure S1 placed the nonpredatory “NR_042149 Fibrobacter succinogenes” [94][140] clustered with “AB540021 Oligoflexus tunisiensis” [82][128], which is predicted to be predatory [3] but as yet unconfirmed (Nakai, pers. comm.). These inconsistencies may be a hint for hunting novel predatory bacteria. Another notable irregularity is “CP075895 Chitinophagaceae bacterium” at 9:10 in Figure 1 and 9:05 in Figure S1. This bacterium, Ca. Cyanoraptor togatus LGM1 [22][61], is the only known obligatory predator within the phylum Bacteroidota and is the only known endobiotic invader outside the Bdellovibrionota (orange)–Pseudomonadota (pale pink) cluster from a little before 3:00 to about 5:25. It is also the first obligatory, intracellular predator of cyanobacteria. Only some of the predatory species of the phyla Bdelovibrionota and Myxococcota are shown in Figure 1 and Figure S1, which would have exhibited more significant proportions of the phyla if all the predatory species were included. However, the importance of the phyla in the phylogeny of predatory bacteria is already explicit in the current Figure 1 and Figure S1 with only selected species.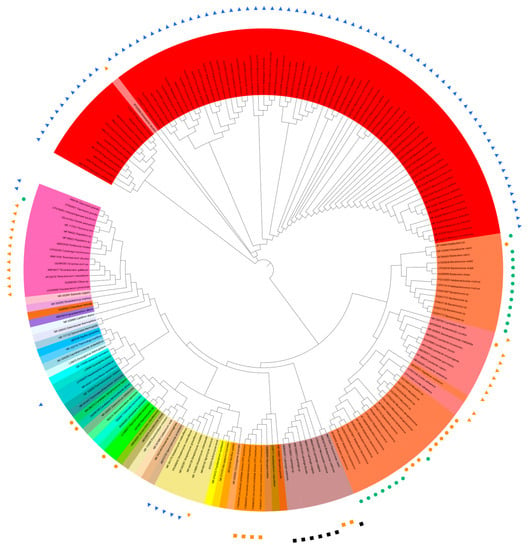

Figure 1. Trans-phylum phylogenetic tree of 135 sequences of predatory bacterial 16S rRNA genes listed [4][12][15][16][17][18][19][20][21][22][23][24][25][26][27][28][29][30][31][32][33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64][65][66][67][68][69][70][71][72][73][74] in Table S1 [4,18,20,31,37,42,47,49,50,61,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120], except [12][37] and 35 sequences from representative, current, and former bacterial phyla listed [75][76][77][78][79][80][81][82][83][84][85][86][87][88][89][90][91][92][93][94][95][96][97][98][99][100][101][102][103][104][105][106][107][108] in Table S2 [121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154]. Predation properties are indicated by the symbols as follows: ●, obligate, epibiotic; ●, obligate, endobiotic or direct invasion; ▲, opportunistic, epibiotic; ▲, opportunistic, group attack; ■, facultative, epibiotic; and, ■, facultative, unknown.
