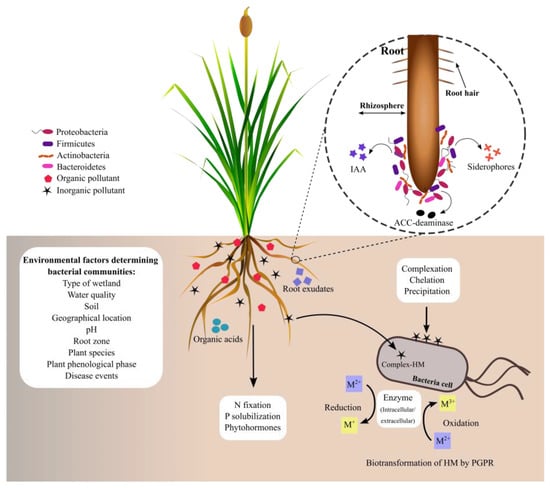
1. Bacteria Associated with the Rhizosphere of Typha spp.
The
Typha genus has a root zone rich in dissolved oxygen and organic carbon that provides favorable conditions for colonization by microorganisms
[1][97]. Plants’ root exudates are essential in the selection and abundance of bacterial communities at the rhizoplane and endosphere roots
[2][98]. Moreover, the composition and structure of bacterial communities are determined by environmental factors, including the type of wetland (natural or artificial), water quality, soil composition, pH, geographical location, root zone, plant species, plant phenological phase, stress, and disease events
[3][4][99,100]. So, rhizospheric bacterial communities can vary in the diversity and abundance of species across the plants, even between plants belonging to the same species. In all ecosystems, bacterial communities are adapted to the plant rhizosphere conditions, where they establish a range of beneficial and deleterious interactions among themselves. Moreover, these bacterial populations contribute to development, growth, and plant adaptation to ecosystems (
Figure 1).
Figure 1. Bacterial phyla commonly associated with the plant rhizosphere. Bacterial phyla comprise species adapted to the rhizosphere conditions, where they interact among themselves by establishing complex interaction networks that contribute to the plant’s fitness to the environmental conditions. Biochemical activities of bacteria are discussed below.
2. Bacterial Communities Associated with Typha Roots in Natural Environments
A few studies have been explored bacterial communities associated with
Typha roots growing in natural environments. Bacteria have been identified by traditional microbiology techniques or by 16S rRNA gene sequencing
(Supplementary Materials Table S1).
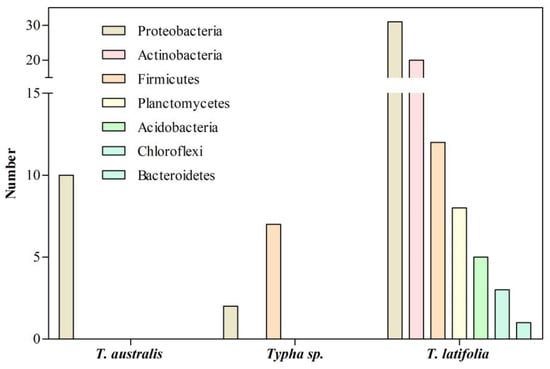
Jha and Kumar
[5][101] isolated ten endophytic diazotrophic bacteria from
T. australis roots collected from ditches of the agricultural farms at Banaras Hindu University (
Figure 2). Bacteria were characterized according to their plant growth-promoting abilities, with
Klebsiella oxytoca GR-3 being the most efficient because of its nitrogenase activity, IAA production, and phosphate solubilization. A plant–bacteria interaction assay showed that
K. oxytoca GR-3 promotes the growth of rice seedlings by increasing the root and shoot length, fresh weight, and chlorophyll content
[5][101]. Thus, root-endophytic
K. oxytoca GR-3 was the first PGPR isolated from the
Typha genus. Despite its plant growth-promoting potential, the effect of
K. oxytoca GR-3 in its host,
T. australis, remains unknown.
Figure 2.
Bacteria associated with the roots of
Typha
species growing in natural environments.
Likewise, Ashkan and Bleakley
[6][102] isolated nine cultivable endophytic bacteria from the roots of
Typha sp. collected at streams of Hot Springs, South Dakota. Then, 16S rRNA sequencing identified seven
Bacillus sp. (
Firmicutes) and two
Pseudomonas sp. (
Proteobacteria) (
Figure 2). These studies were limited because the screening was focused on cultivable bacteria. Furthermore, the plant growth-promoting abilities of the isolates were not tested, so their role in
Typha sp. is unknown.
On the other hand, PCR-guided analysis of the 16S rRNA gene was performed to elucidate the composition of the root-associated bacterial microbiota of
T. latifolia growing in a wetland in the natural paradise “Le Mortine Oasis”, Campania, southern Italy
[7][65]. The root-associated microbiota of
T. latifolia was dominated by the phylum
Proteobacteria (31 isolates, 39 %), followed by
Actinobacteria (20 isolates, 25%),
Firmicutes (12 isolates, 15%),
Planctomycetes (8 isolates, 10%),
Acidobacteria (5 isolates, 6%), and
Chloroflexi (3 isolates, 4%) (
Figure 2). Among them,
Proteobacteria dominate rhizoplane, rhizosphere, and at least a 2 m radius of surrounding water of
T. latifolia collected in the wetland. These results agree with previous data obtained for the plant microbiota, mainly dominated by
Proteobacteria [8][103].
Rhizobiales,
Rhodobacterales, and
Pseudomonadales were the most abundant
Proteobacteria associated with
T. latifolia roots
[7][65]. This is due to
Proteobacteria being fast-growing r-strategists that are able to utilize a broad range of root-derived carbon substrates, showing adaptation to the diverse plant rhizospheres
[8][9][103,104]. The most abundant phyla,
Proteobacteria,
Actinobacteria, and
Firmicutes, include bacterial species of the families
Rhizobiaceae,
Pseudomonadaceae,
Streptomycetaceae, and
Bacillaceae that potentially promote plant growth
[8][103]. These bacterial species could be involved in adapting
T. latifolia to the environmental conditions in the “Le Mortine Oasis”. These results indicated that sequencing analysis generated a broader overview of bacteria associated with plant roots and is an excellent strategy for exploring bacterial diversity and abundance in the plant rhizosphere.
Cultivable bacteria from
T. latifolia rhizoplane were also isolated, and determined their biofilm formation capacity
[7][65]. The best biofilm-forming bacteria were identified by 16S rRNA gene sequencing, revealing the presence of
Microbacterium chocolatum,
Streptomyces sp.,
Streptomyces mirabilis,
Rhodococcus sp.,
Wautersiella sp.,
Pseudomonas sp.,
Janthinobacterium lividum, and family members of
Xanthomonadaceae,
Enterobacteriaceae, and
Bacillaceae, which colonize the
T. latifolia rhizoplane. However, the PGPR activities of bacterial isolates were not determined, as they could contribute to the adaptation and growth of
T. latifolia in the “Le Mortine Oasis”
[7][65]. Overall, combining 16S rRNA gene sequencing and isolation by culture-dependent techniques offers a broader view of microbial populations associated with
T. latifolia roots
(Supplementary Materials Table S1). Even bacterial isolates or consortia could be tested in interaction assays with their host to identify the most efficient isolates that promote the growth of
T. latifolia.
3. Bacterial Communities Associated with the Roots of Typha Exposed to Heavy Metals
Typha species have been used in phytoremediation because of their abilities to remove
heavy metals (HMs)HMs from the surrounding environment
[10][11][12][1,53,66]. It has been demonstrated that HM removal is enhanced by microorganisms associated with the plant roots
[13][105]. So, identifying bacterial populations associated with the roots of
Typha species has gained importance (
Table 1). However, available information has been obtained from
Typha species collected in different environmental conditions and using either culture-dependent or gene sequencing strategies (
Figure 3). So, the information on specific bacterial communities of each
Typha species is scarce for defining the
Typha microbiome.
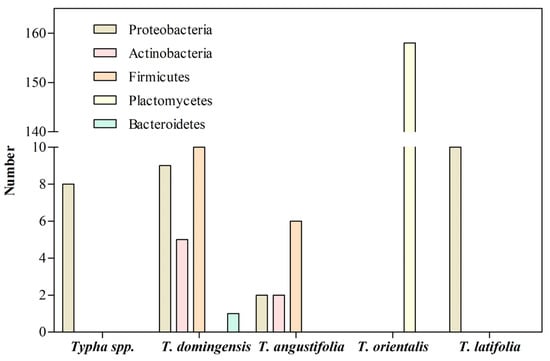
Figure 3.
Bacteria associated with the roots of
Typha
species growing in presence of HMs.
The first study with a
Typha species was conducted by Pacheco-Aguilar et al.
[14][69], who isolated bacteria that colonize the roots of
Typha sp. growing in an artificial wetland to treat the wastewater of a tannery contaminated with high concentrations of organic matter, chromium, nitrogen, and sulfur compounds. Eight bacterial strains of the phylum
Proteobacteria were isolated, including three
Pseudomonas sp., two
Acinetobacter sp., two
Alcaligenes sp., and one
Ochrobactrum sp. (
Table 13; Supplementary Materials Table S2). The characteristics of the wastewater agree with the predominance of bacteria belonging to the phylum
Proteobacteria, which could be involved in degrading organic matter.
It is known that the versatile
Pseudomonas genus can tolerate HMs because of its ability to adapt to various environmental conditions
[18][19][108,109].
Ochrobactrum strains have been shown to alleviate Cd toxicity in spinach (
Spinacia oleracea L.)
[20][110] and rice (
Oryza sativa)
[21][111] and adsorb Cd and Cr in their cell wall due to exopolysaccharides content
[22][23][24][112,113,114]. Similarly, some
Acinetobacter strains have been applied to improve the removal efficiency of heavy metals such as Cr
[25][26][27][115,116,117], while
Alcaligenes has been reported to be tolerant to Cd, Cu, Ni, Zn, and Cr
[28][29][118,119].
It is probable that Pseudomonas sp., Acinetobacter sp., Alcaligenes sp., and Ochrobactrum sp. play an essential role in adapting Typha sp. to wetland conditions and improving the Cr removal by the plant.
Shehzadi et al.
[15][106] isolated cultivable endophytic bacteria from the shoots and roots of
T. domingensis grown in wetlands to treat textile effluents contaminated with organic matter, phosphates, sulphates, nitrates, and Ni, Fe, Cr, and Cd. These endophytic bacteria belong to the phyla
Proteobacteria,
Firmicutes,
Actinobacteria, and
Bacteroidetes (
Table 1). Among them,
Bacillus sp. TYSI17,
Microbacterium arborescens TYSI04,
Microbacterium sp. TYSI08,
Rhizobium sp. TYRI06,
Pantoea sp. TYRI15, and
Pseudomonas fluorescens TYSI35 exerted PGPR activities and exhibited the best textile effluent degrading activity. These results suggest that bacterial strains could be involved in the growth and adaptation of
T. domingensis to the wetland conditions. The most efficient bacterial strains could be used to remediate industrial effluents.
Saha et al.
[16][64] isolated ten cultivable endophytic bacteria from the roots of
T. angustifolia grown at a uranium mine tailing contaminated with iron. The 16S rRNA gene sequencing of the isolates indicated that the endophytic bacteria belonged to the phyla
Firmicutes,
Proteobacteria, and
Actinobacteria (
Table 1). All bacterial isolates exerted biochemical activities, including auxin synthesis, siderophore production, and nitrogen fix. Plant–bacteria interaction assays showed that a consortium including ten isolates promotes the growth of
T. angustifolia and rice seedlings, indicating that isolates are PGPR, which could participate in the adaptation of
T. angustifolia to the contaminated site. Thus, the consortium has the biotechnological potential to be applied for plant-bacteria remediation purposes.
Additionally, Zhou et al.
[17][107] investigated the composition and abundance of ammonium-oxidizing bacteria in the rhizosphere of
T. orientalis exposed to Cu, Cr, Pb, Cd, and Zn on the shores of Jinshan Lake Park in China, and 16S rRNA sequencing revealed that the bacterial communities belonged to the phylum
Planctomycetes (
Table 1). The bacterial composition of the rhizosphere was affected by the presence of Cu, Pb, and Zn, the availability of nitrogen, and even the stage of growth and the year’s season. The phylum
Planctomycetes has been associated with soil microbial communities in response to stress from heavy metals, such as Hg, Cd, and Cr
[30][31][120,121]. This phylum can detoxify heavy metals by secreting extracellular substances, such as polysaccharides and proteins, reducing the toxic effects of metals
[32][33][122,123]. The increase in the
Planctomycetes population has also been related to an acidic pH, and this phylum’s diversity varies according to environmental conditions
[30][31][120,121].
Rolón-Cárdenas et al.
[11][53] isolated four endophytic strains of
Pseudomonas rhodesiae from the roots of
T. latifolia exposed to Cd in a contaminated site. Bacterial strains showed high tolerance to Cd and exerted biochemical activities such as PGPR (
Table 13; Supplementary Materials Table S2). Plant–bacteria interaction assays showed that
P. rhodesiae strains promote the growth of
Arabidopsis thaliana seedlings exposed and non-exposed to Cd. Likewise,
P. rhodesiae decreases oxidative stress in
T. latifolia seedlings exposed to Cd and improves Cd translocation to the shoot in an axenic hydroponic system
[34][124]. Additionally, Rubio-Santiago et al.
[35][125] isolated endophytic bacteria
P. azotoformans,
P. fluorescens,
P. gessardii, and
P. veronii from the roots of
T. latifolia growing at a Pb- and Cd-contaminated site. Bacterial strains exerted biochemical activities such as PGPR and promoted the growth of
T. latifolia seedlings exposed and non-exposed to either Pb or Cd. Thus, these bacterial strains could be used in plant–bacteria interactions for phytoremediation with
T. latifolia.
The studies of microorganisms associated with the roots of the Typha genus were focused on isolating PGPR tolerant to HMs, because they promote plant growth and improve phytoextraction capacity in contaminated environments. However, the mechanisms by which endophytic bacteria enhance phytoremediation in Typha remain unclear. Furthermore, no Typha-bacteria model is available to determine the global mechanisms involved in phytoremediation.
4. Other Microorganisms Associated with the Rhizosphere of Typha spp.
The rhizosphere is the habitat of many microorganisms and invertebrates, considered one of Earth’s most dynamic interfaces
[9][104]. Although bacteria are the main inhabitants of the rhizosphere, the plant roots can be colonized by fungi, protozoa, rotifers, nematodes, and microarthropods, which are attracted by rhizodeposits, nutrients, exudates, border cells, and mucilage released by the plant root
[4][100].
The presence of endophytic fungi has been reported in
T. latifolia roots, including
Aspergillus sp.,
Myrothecium sp.,
Phoma sp.,
Penicillium sp.,
Acremonium sp., and
Fusarium sp., while in its rhizome
Penicillium sp.,
Myrothecium sp., and
Fusarium sp. have been found
[36][173]. Furthermore, the efficiency of
Aspergillus niger,
Acremonium sp.,
Aureobasidium sp.,
Cephalosporium sp., and
Fusarium sp., to degrade pollutants or absorb HMs has been demonstrated; thus, they are an option in mycoremediation
[36][37][173,174]. On the other hand, a study conducted in Zhejiang Province, Eastern China, by Guan et al.
[38][175] reported the presence of common fungal species including
Pleosporales sp.,
Teratosphaeria microspora,
Geotrichum candidum, Engyodontium album, Blastocladiales sp., uncultured soil fungus,
Fusarium graminearum, and
Cladosporium bruhnei in the rhizosphere of
T. latifolia and
T. orientalis. While
Paramicrosporidium saccamoebae, Rhynchosporium secalis,
Acremonium roseolum, and
Cladosporium sphaerospermun were found in
T. latifolia, and uncultured soil fungus in
T. orientalis.
Finally, arbuscular vesicular (AV) fungi are known to increase plant resistance to heavy metals, and this differs according to fungal species, plant, and environmental conditions
[39][176].