Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Lindsay Dong and Version 1 by Alexander Yuile.
The CDKN2A and CDKN2B genes are located on the short arm of chromosome 9. CDKN2A encodes for two proteins, p14 and p16, and CDKN2B encodes for p15. These proteins regulate cell growth and angiogenesis. Interpreting the impact of CDKN2A/B alterations on astrocytoma prognosis is complicated by rthecent changes in tumour classification and a lack of uniform standards for testing CDKN2A/B. While the prognostic impact of CDKN2A/B HD is established, the role of different CDKN2A/B alterations—heterozygous deletions (HeD), point mutations, and promoter methylation—is less clear. Consequently, how these alternations should be incorporated into patient management remains controversial.
- CDKN2A/B alterations
- CDKN2A/B homozygous deletions
- temozolomide
1. Introduction
In 2016, the WHO Classification of Tumours of the Central Nervous System (revised 4th edition) incorporated isocitrate dehydrogenase 1 and 2 (IDH1/2) mutation status into the classification of diffuse gliomas [1]. IDH1/2 mutations (point mutations in either IDH1 codon R132 or IDH2 codon R172) lead to IDH dysfunction, which converts alpha-ketoglutarate to R-2-hydroxygluterate, driving oncogenesis and global epigenetic changes [1,2,3,4][1][2][3][4]. The WHO Classification in 2016 further stratified diffuse IDH-mutant gliomas into oligodendrogliomas and astrocytomas based on the respective presence or absence of chromosome 1p and 19q co-deletions (see Figure 1) [1].
IDH-mutant astrocytomas account for 80% of WHO grades 2 to 3 and 5% of high-grade astrocytomas [5,6][5][6]. When compared to IDH-wildtype glioblastomas, patients with IDH-mutant astrocytomas are younger at diagnosis (30–40 years vs. over 50 years). In addition, IDH-mutant astrocytomas have a more favourable prognosis compared to IDH-wildtype glioblastomas, even in high-grade cases, with grade 4 IDH-mutant astrocytomas having a median overall survival (OS) of 31 months, compared to IDH-wildtype glioblastomas with a median OS of 13 months [5,6][5][6]. Unfortunately, a proportion of IDH-mutant astrocytomas have poor outcomes similar to those of IDH-wildtype glioblastomas [7].
CDKN2A/B HD are identified in approximately 22% of IDH-mutant astrocytomas [8] and are thought to lead to the loss of cell cycle control and promote cell proliferation [9].
Due to the improved prognosis of tumours previously classified as IDH-mutant glioblastomas compared to IDH-wildtype glioblastomas, these have been reclassified as astrocytoma, IDH-mutant, CNS WHO grade 4 in the fifth edition of the WHO Classification of Tumours of the Central Nervous System (WHO CNS5, 2021). A hallmark of WHO CNS5 is the integration of molecular markers into tumour grading. As such, IDH-mutant astrocytomas with CDKN2A/B HD are classified as grade 4 tumours independent of morphologic features. Therefore, a diagnosis of astrocytoma, IDH mutant, or CNS WHO grade 4 requires either morphologic features of a glioblastoma, namely necrosis or microvascular proliferation, or homozygous deletion of CDKN2A and/or CDKN2B (see Figure 1) [1,14][1][10].
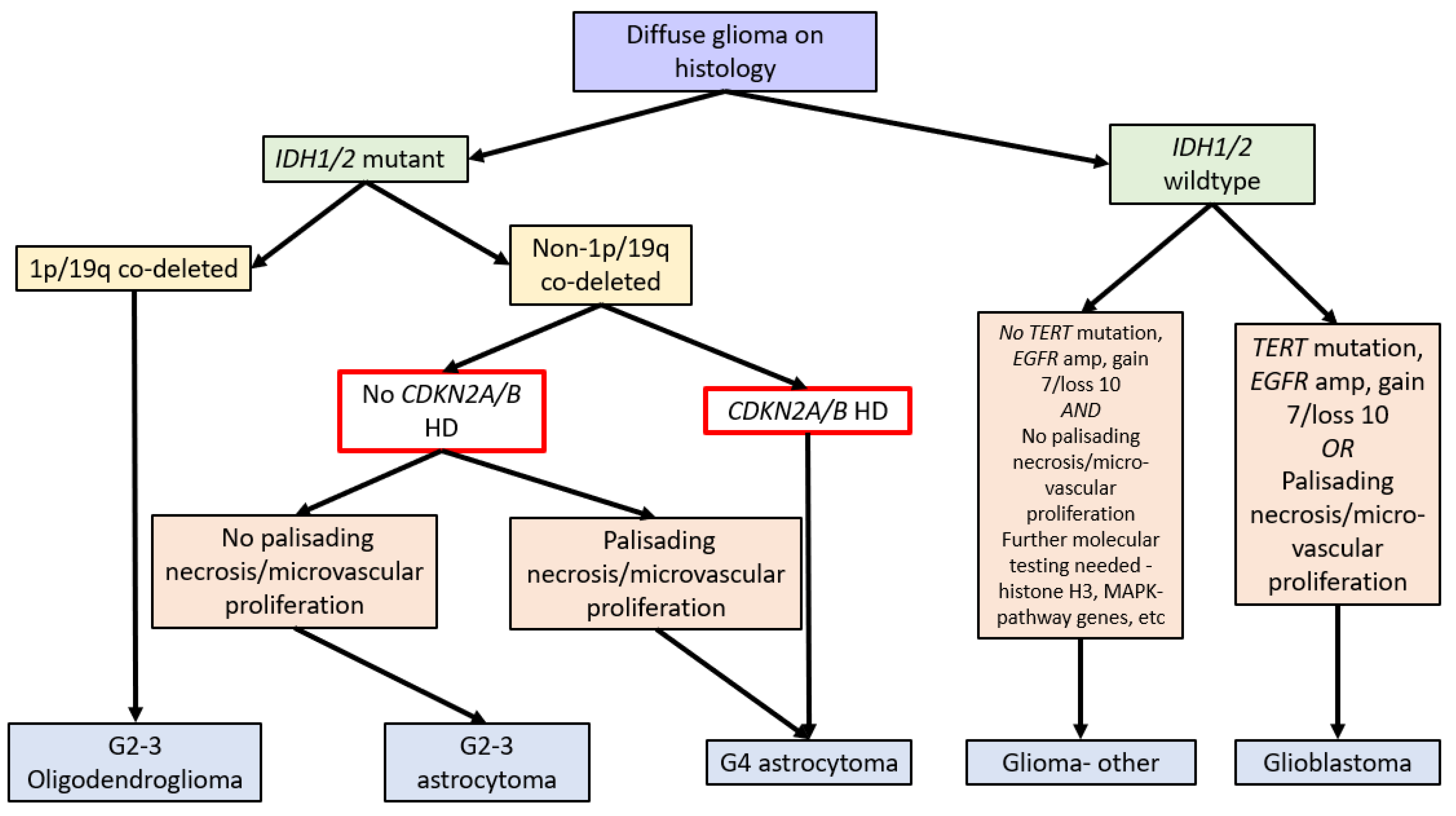 The significance of CDKN2A/B alterations in gliomas is difficult to assess in historical cohorts. Prior to 2016, many studies classified tumours based only on morphology. Consequently, tumours previously classified as astrocytomas on morphological grounds are likely to include tumours that are currently classified as astrocytoma, IDH-mutant, oligodendroglioma IDH-mutant, 1p/19q codeleted or glioblastoma, and IDH-wildtype [15,16,17][11][12][13]. Interpretation of the CDKN2A/B literature is further complicated by the multiple different techniques used to interrogate the CDKN2A and CDKN2B genes and whether one or both genes are interrogated. In addition, there is ambiguity concerning the significance of isolated CDKN2A or CDKN2B loss compared to loss of both CDKN2A and CDKN2B [11,13,18,19,20][14][15][16][17][18].
The significance of CDKN2A/B alterations in gliomas is difficult to assess in historical cohorts. Prior to 2016, many studies classified tumours based only on morphology. Consequently, tumours previously classified as astrocytomas on morphological grounds are likely to include tumours that are currently classified as astrocytoma, IDH-mutant, oligodendroglioma IDH-mutant, 1p/19q codeleted or glioblastoma, and IDH-wildtype [15,16,17][11][12][13]. Interpretation of the CDKN2A/B literature is further complicated by the multiple different techniques used to interrogate the CDKN2A and CDKN2B genes and whether one or both genes are interrogated. In addition, there is ambiguity concerning the significance of isolated CDKN2A or CDKN2B loss compared to loss of both CDKN2A and CDKN2B [11,13,18,19,20][14][15][16][17][18].

2. Normal Role of CDKN2A/B and Effect of Their Deletion
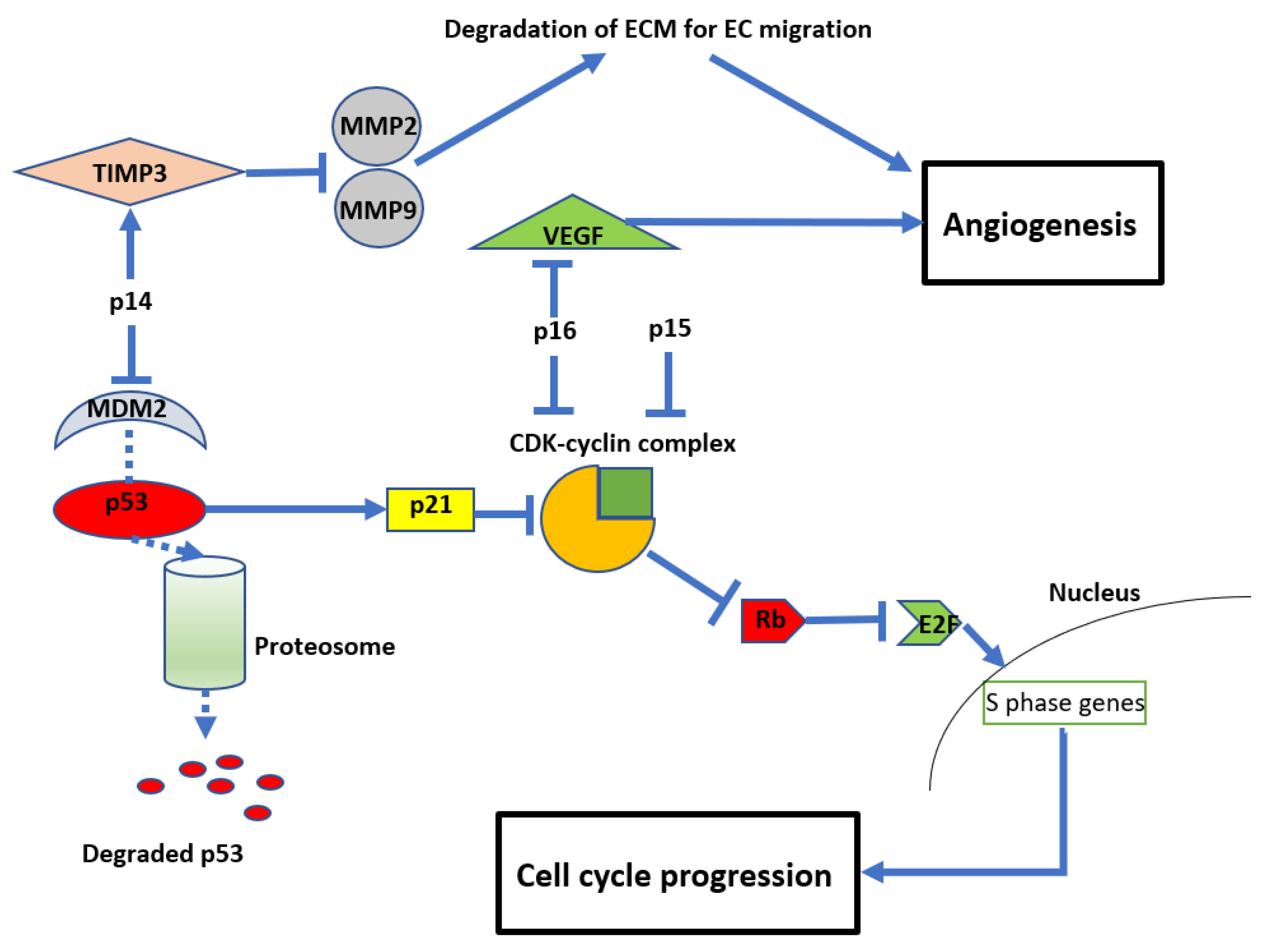
CDKN2A and CDKN2B are adjacent to each other on chromosome 9p21, with CDKN2B being 25 kilobases (kb) centromeric to CDKN2A [34][19]. These genes code for three proteins that suppress the oncogenic cyclin-dependent kinase (CDK) pathway. CDKN2A encodes for p14 and p16, and CDKN2B encodes for p15 [35][20]. Early cytogenetic studies identified recurrent loss of the short arm of chromosome 9 in glioma cell lines [21[21][22][23][24],22,23,36], many involving the 9p21 locus [22[22][25],37], which includes CDKN2A and CDKN2B [23,36][23][24]. The resultant loss of p14, p15, and p16 proteins from CDKN2A/B HD leads to dysregulation of the cell cycle and other parallel oncogenic processes (see Figure 2). Reflecting this, inactivation of CDKN2A function has been reported in a variety of other malignancies, including breast cancer, lung cancer, head and neck cancer, melanoma, and bladder cancer [9]. In normal cells, the retinoblastoma (Rb) protein prevents cell growth by binding to the transcription factor E2F, preventing its translocation into the nucleus. A complex formed by cyclin D and CDK4/6 can phosphorylate Rb, thereby releasing E2F and allowing translocation into the nucleus, leading to cell growth. The products of CDKN2A/B, p15 and p16, can directly inhibit the formation of the CDK4/6-cyclin D complex, maintaining the association between E2F and Rb [39,40,41][26][27][28] and preventing cell cycle progression. Another product of CDKN2A is p14, which acts on cyclin-CDK complexes indirectly by inhibiting MDM2. MDM2 tags p53, targeting it for ubiquitination and subsequent proteasomal degradation. 14 prevents MDM2 tagging, resulting in p53 stabilisation. This in turn promotes the cellular accumulation of the inhibitory protein p21, which blocks several cyclin-CDK complexes and promotes cell cycle arrest [42,43][29][30]. Due to these important functions of the protein products of CDKN2A/B, their deletion enhances oncogenic potential and leads to unregulated cellular proliferation (see Figure 32) [42,43][29][30].
Figure 32. Diagram showing the anti-proliferative and anti-angiogenic effects of CDKN2A/B Mouse double minute 2 homolog (MDM2), tissue inhibitor of metalloproteinase 3 (TIMP3), matrix metalloproteinases (MMP).
In addition to their role in regulating cell growth, CDKN2A/B also impact angiogenesis (see Figure 32). For example, p14 (unrelated to its inhibition of MDM2) can also inhibit endothelial cell migration required for angiogenesis by stimulating the expression of tissue inhibitor of metalloproteinase 3 (TIMP3), which inhibits matrix metalloproteinases (MMP) 2 and 9 [44][31]. MMPs are required to degrade the extracellular matrix to allow endothelial cell migration and subsequent vessel formation [45,46][32][33].
3. Identification of CDKN2A/B Deletions
A variety of methods can be used to evaluate CDKN2A/B HD. These include single-nucleotide polymorphism (SNP) microarrays [31[34][35],53], next-generation sequencing (NGS) [12[36][37],29], DNA-based methylation studies [11,13[14][15][16][17][18],18,19,20], and fluorescent in situ hybridisation (FISH) [10,28][38][39]. It should be noted that the accuracy of this variety of methods depends on the specific assay types used, as the genomic/cytogenetic resolution of each method differs.
Although SNP arrays, NGS, and methylation arrays possess greater resolution for individual gene-level detection, many studies combine CDKN2A and CDKN2B in the assessment of HD [11,18,29,31,53][14][16][34][35][37]. The accuracy of these methods is determined by the degree and depth of coverage of the genes of interest. NGS methods used in the literature to date include targeted gene panels [12][36] and whole exome sequencing (WES) [29][37], whereas methylation arrays include a combination of the HumanMethylation450 (450k) and MethylationEPIC (850k) arrays (Illumina, San Diego, CA, USA) [11,13,18,19,20][14][15][16][17][18]
Fluorescence in situ hybridization (FISH) can be used to detect deletions and has been validated against methods utilising polymerase chain reaction (PCR) [54][40]. Thresholds of detection for FISH need to be around 20% to 30% tumour cells with HD [10,55][38][41]. A commonly used FISH probe in clinical diagnostic practise, the Vysis CDKN2A/CEP 9 FISH Probe Kit (Abbott Laboratories, North Chicago, IL, USA), is large and spans CDKN2A, CDKN2B, and MTAP genes [56][42].
Immunohistochemistry (IHC) has been used to identify CDKN2A HD in gliomas with mixed results. Given the close proximity of the CDKN2A and MTAP genes (see Figure 2), loss of MTAP immunoreactivity has been suggested as a surrogate for CDKN2A HD [32][43] and has been demonstrated in mesothelioma [57,58][44][45].
However, Satomi et al. did show that loss of p16 immunoreactivity correlated with clinical outcome in IDH-mutant astrocytomas [32][43]. While this is supported by other studies that demonstrated p16-negative tumours on IHC had a high negative predictive value for CDKN2A HD in adult and paediatric morphologic glioblastomas [59][46], other studies reported p16/CDKN2A discordance with the IHC method [28][39]. Sensitivity and specificity for p16 immunoreactivity in detecting CDKN2A HD have been reported as 78–94% and 70–82%, respectively [32][43].
4. CDKN2A/B Deletions in Clinical Studies
4.1. Initial Clinical Studies
Initial studies by Schmidt et al. [60][47] and Giani and Finocchiaro et al. [61][48] confirmed that CDKN2A HD was present in patients’ tumours and not just in glioma cell lines but did not assess CDKN2B. Giani and Finocchiaro et al. demonstrated CDKN2A HD in over 30% of gliomas (not further defined) and CDKN2A HeD in 25% [61][48]. Moulton et al. analysed 27 glioblastomas (not further defined) and identified 9 with CDKN2A HD, 3 with a heterozygous deletion, and one with a point mutation [62][49].4.2. Clinical Outcomes of CDKN2A/B Deletion in the Pre-Molecular Classification Era (Pre-2016 WHO CNS Tumour Classification)
4.2.1. Correlation with High- and Low-Grade Gliomas
Initial studies described the relationship between CDKN2A/B and biologic markers of tumour aggressiveness (tumour grade and Ki-67 index). Sonoda et al. suggested CDKN2A/B deletions may have a role in gliomagenesis and therefore more aggressive tumour biology. Using single-strand conformation polymorphism (SSCP) and quantitative polymerase chain reaction (qPCR), they showed an increased incidence of CDKN2A/B HD in high-grade gliomas (44%, n = 12/27) compared to low-grade gliomas (10%, n = 1/10) [26][50]. Building on this concept, Ono et al. (1996) used multiplex PCR to assess CDKN2A/B HD in 50 astrocytomas and found a positive correlation between the Ki-67 index and CDKN2A HD (5/20 grade 3 astrocytomas and 6/13 glioblastomas had CDKN2A HD). CDKN2A HD was not identified in 17 grade 2 astrocytomas [64][51].4.2.2. Correlation with Survival
In 2006, Dehais et al. reported that CDKN2A HD was a negative prognostic factor in a heterogeneous group of gliomas that included anaplastic astrocytomas, oligoastrocytomas, and oligodendrogliomas. Although 1p/19q status was assessed, the authors did not identify which cases had CDKN2A HD and 1p/19q co-deletion [15][11]. However, other reports did not find an association between CKDN2A HD and clinical outcome [66,67][52][53]. This may reflect differences in methodology and/or patient selection for tumours classified by morphology alone. One of these studies (Rich et al.) used a DNA microarray to assess the prognostic impact of CDKN2A deletion in patients older than 50 years. Although IDH status was not reported in the study, this population was likely enriched for IDH-wildtype tumours, and it was later shown that CDKN2A deletions lack prognostic impact in these tumours [66][52].4.3. Clinical Outcomes in the Post-Molecular Classification Era (Post-2016 WHO CNS Tumour Classification)
4.3.1. Incorporation of CDKN2A/B Status into the fifth Edition of the WHO Classification (2021)
In 2020, the Consortium to Inform Molecular and Practical Approaches to CNS Tumour Taxonomy (cIMPACT-NOW), upgrade 5, published recommendations for grading criteria and terminologies in IDH-mutant astrocytomas. After reviewing the literature on multiple potential prognostic biomarkers, including CDKN2A/B HD, other Rb pathway genes, PIK3R1 and PIK3CA mutations, PDGFRA and MYCN amplification, reduced global DNA methylation, genomic instability (high copy number variants or somatic mutations), and mitotic activity and proliferation indices, they concluded that while “significant mitotic activity” should remain as a criterion for distinguishing grade 3 from grade 2 IDH-mutant astrocytomas, if CDKN2A/B HD, necrosis, or microvascular proliferation was present, a grade 4 designation was appropriate [7].4.3.2. Literature That Supports CDKN2A/B Stratification
A prime example of supporting literature is Reis et al who were one of the earliest to report on the prognostic impact of CDKN2A HD in the setting of IDH mutations. They identified CDKN2A deletions as a prognostic marker specifically in IDH-mutant grade 2 and 3 gliomas. The authors analysed 270 gliomas and identified CDKN2A deletions via FISH in 57/108 grade 2 astrocytomas, 31/61 grade 3 astrocytomas, 23/96 oligodendrogliomas, and 19/49 oligoastrocytomas, inclusive of both homozygous and heterozygous CDKN2A deletions. The authors assessed tumours for 1p/19q deletion if they were not morphologic astrocytomas and assessed all tumours for IDH1/2 mutations by genome sequencing. They reported worse overall survival in grade 2 and 3 gliomas after adjusting for age, sex, and IDH mutation (HR 1.6, 95% CI = 1.0–2.4, p = 0.03). This significance was maintained in the astrocytoma subgroup (HR 2.0, 95% CI 1.1–3.5, p = 0.02) but not for oligodendrogliomas or oligoastrocytomas (HR 0.7, 95% CI 0.2–2.0, p = 0.5 and HR 0.8, 95% CI 0.3–2.4, p = 0.7, respectively). Again, a portion of these morphologic oligodendrogliomas in this cohort would no longer be classified as such without the corresponding molecularly confirmed 1p19q co-deletion. Interestingly, the presence of deletions in the IDH-mutant/ATRX expression loss astrocytoma group, without TP53 mutation, was non-prognostic (p = 0.2) [68][54]. Furthermore, as ATRX loss and TP53 mutations are strongly associated with IDH-mutant astrocytomas, it is unclear what this ATRX/TP53 discordance represents in IDH-mutant gliomas. Interestingly, given the FISH probe used covers a broad genomic region at 9p21, CDKN2B status can be said to be assessed by proxy.4.3.3. Literature That Counters CDKN2A/B Stratification
Not all studies supported the use of CDKN2A/B in IDH-mutant astrocytomas. One such example is Roy et al. who analysed the 9p region lost in malignancies by analysing two cohorts (the first group being 10,985 samples from 33 different cancer types and the second group being 540 low-grade gliomas from three databases) and reported that CDKN2A inactivation did not promote tumour aggressiveness. Even when accounting for IDH and 1p/19q status (IDH-mutant 1p/19q non-deleted astrocytoma), there was no survival impact of CKDN2A HD. While they did show that heterozygous loss was associated with poor OS, mRNA expression was not altered. It was therefore postulated that this survival impact was due to the loss of other 9p genes [30][55]. It is unclear why this report differs from the majority of other studies, but it highlights that not all studies support the role of CDKN2A/B HD as a prognostic marker in IDH-mutant astrocytomas.5. Management of Tumours with CDKN2A/B Homozygous Deletions
There is no clear consensus on the treatment of IDH-mutant astrocytomas with CDKN2A/B HD, and reports related to their management are scarce. Reflecting this ambiguity, the current joint American Society of Clinical Oncology and Society of Neuro-Oncology guidelines recommend grade 4 astrocytomas be treated with concurrent temozolomide-radiotherapy with sequential temozolomide or radiotherapy alone with sequential temozolomide [70][56]. However, given the evidence that CDKN2A/B HD alters tumour biology (increased angiogenesis and cell growth), it cannot assume that these tumours will be as susceptible to temozolomide as their non-deleted counterparts. Unfortunately, the evidence for treatment specifically for CDKN2A/B HD astrocytomas is minimal. In 2000, Iwadate et al. investigated the relationship between CDKN2A deletion, p16 expression, and chemosensitivity to 30 different cytotoxic agents in vitro. They analysed 56 astrocytoma specimens (based on morphologic criteria, IDH status unknown) and found 17 specimens had p16 alterations (CDKN2A HD = 7, CDKN2A mutation = 5, p16 loss on IHC = 5). When looking at samples with p16 alterations, they found that deletions correlated with increased sensitivity to anti-metabolite agents but not to alkylating agents, antibiotics, topoisomerase inhibitors, or anti-microtubule agents [71][57].6. Conclusions
CDKN2A/B HD have a direct oncogenic effect through loss of cell cycle inhibition and other parallel processes and are a molecular marker that influences grading and survival in IDH-mutant astrocytomas. Overall, the evidence supports the use of CDKN2A/B HD as a negative prognostic marker in IDH-mutant astrocytomas. However, there is a significant variation in certainty, methods used for deletion detection, and the quality of the presented literature. There are also inaccuracies resulting from misclassification of tumours in older studies based on the revised WHO classification. These limitations hamper conclusions regarding the certainty and depth of impact CDKN2A/B HD has on prognosis and management and how this impact is affected by other co-occurring molecular alterations. Therefore, the strongest evidence for CDKN2A/B HD in IDH-mutant astrocytomas must come from prospective reports with the current WHO 2021 classification.
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820.
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009, 462, 739–744.
- Huang, L.E. Friend or foe-IDH1 mutations in glioma 10 years on. Carcinogenesis 2019, 40, 1299–1307.
- Yuile, A.; Satgunaseelan, L.; Wei, J.; Kastelan, M.; Back, M.F.; Lee, M.; Wei, H.; Buckland, M.E.; Lee, A.; Wheeler, H.R. Implications of Concurrent IDH1 and IDH2 Mutations on Survival in Glioma—A Case Report and Systematic Review. Curr. Issues Mol. Biol. 2022, 44, 5117–5125.
- Eckel-Passow, J.E.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Sicotte, H.; Pekmezci, M.; Rice, T.; Kosel, M.L.; Smirnov, I.V.; et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N. Engl. J. Med. 2015, 372, 2499–2508.
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765–773.
- Brat, D.J.; Aldape, K.; Colman, H.; Figrarella-Branger, D.; Fuller, G.N.; Giannini, C.; Holland, E.C.; Jenkins, R.B.; Kleinschmidt-DeMasters, B.; Komori, T.; et al. cIMPACT-NOW update 5: Recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020, 139, 603–608.
- Lu, V.M.; O’Connor, K.P.; Shah, A.H.; Eichberg, D.G.; Luther, E.M.; Komotar, R.J.; Ivan, M.E. The prognostic significance of CDKN2A homozygous deletion in IDH-mutant lower-grade glioma and glioblastoma: A systematic review of the contemporary literature. J. Neurooncol. 2020, 148, 221–229.
- Sharpless, N.E. INK4a/ARF: A multifunctional tumor suppressor locus. Mutat. Res. 2005, 576, 22–38.
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251.
- Dehais, C.; Laigle-Donadey, F.; Marie, Y.; Kujas, M.; Lejeune, J.; Benouaich-Amiel, A.; Pedretti, M.; Polivka, M.; Xuan, K.-H.; Thillet, J.; et al. Prognostic stratification of patients with anaplastic gliomas according to genetic profile. Cancer 2006, 107, 1891–1897.
- James, C.D.; Galanis, E.; Frederick, L.; Kimmel, D.W.; Cunningham, J.M.; Atherton-Skaff, P.J.; O’Fallon, J.R.; Jenkins, R.B.; Buckner, J.C.; Hunter, S.B.; et al. Tumor suppressor gene alterations in malignant gliomas: Histopathological associations and prognostic evaluation. Int. J. Oncol. 1999, 15, 547–600.
- Cairncross, J.G.; Ueki, K.; Zlatescu, M.C.; Lisle, D.K.; Finkelstein, D.M.; Hammond, R.R.; Silver, J.S.; Stark, P.C.; Macdonald, D.R.; Ino, Y.; et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J. Natl. Cancer Inst. 1998, 90, 1473–1479.
- Shirahata, M.; Ono, T.; Stichel, D.; Schrimpf, D.; Reuss, D.E.; Sahm, F.; Koelsche, C.; Wefers, A.; Reinhardt, A.; Huang, K.; et al. Novel, improved grading system(s) for IDH-mutant astrocytic gliomas. Acta Neuropathol. 2018, 136, 153–166.
- Yoda, R.A.; Marxen, T.; Longo, L.; Ene, C.; Wirsching, H.-G.; Keene, C.D.; Holland, E.C.; Cimino, P.J. Mitotic Index Thresholds Do Not Predict Clinical Outcome for IDH-Mutant Astrocytoma. J. Neuropathol. Exp. Neurol. 2019, 78, 1002–1010.
- Korshunov, A.; Casalini, B.; Chavez, L.; Hielscher, T.; Sill, M.; Ryzhova, M.; Sharma, T.; Schrimpf, D.; Stichel, D.; Capper, D.; et al. Integrated molecular characterization of IDH-mutant glioblastomas. Neuropathol. Appl. Neurobiol. 2019, 45, 108–118.
- Cimino, P.J.; Zager, M.; McFerrin, L.; Wirsching, H.-G.; Bolouri, H.; Hentschel, B.; von Deimling, A.; Jones, D.; Reifenberger, G.; Weller, M.; et al. Multidimensional scaling of diffuse gliomas: Application to the 2016 World Health Organization classification system with prognostically relevant molecular subtype discovery. Acta Neuropathol. Commun. 2017, 5, 39.
- Cimino, P.J.; Holland, E.C. Targeted copy number analysis outperforms histologic grading in predicting patient survival for WHO grades II/III IDH-mutant astrocytomas. Neuro Oncol. 2019, 21, 819–821.
- Worsham, M.J.; Chen, K.M.; Tiwari, N.; Pals, G.; Schouten, J.P.; Sethi, S.; Benninger, M.S. Fine-mapping loss of gene architecture at the CDKN2B (p15INK4b), CDKN2A (p14ARF, p16INK4a), and MTAP genes in head and neck squamous cell carcinoma. Arch. Otolaryngol. Head. Neck Surg. 2006, 132, 409–415.
- Cánepa, E.T.; Scassa, M.E.; Ceruti, J.M.; Marazita, M.C.; Carcagno, A.L.; Sirkin, P.F.; Ogara, M.F. INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB Life 2007, 59, 419–426.
- Rey, J.A.; Bello, M.J.; de Campos, J.M.; Kusak, M.E.; Ramos, C.; Benitez, J. Chromosomal patterns in human malignant astrocytomas. Cancer Genet. Cytogenet. 1987, 29, 201–221.
- James, C.D.; He, J.; Carlbom, E.; Nordenskjold, M.; Cavenee, W.K.; Collins, V.P. Chromosome 9 Deletion Mapping Reveals Interferon α and Interferon β-1 Gene Deletions in Human Glial Tumors. Cancer Res. 1991, 51, 1684–1688.
- Serrano, M.; Hannon, G.J.; Beach, D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993, 366, 704–707.
- Nobori, T.; Miura, K.; Wu, D.J.; Lois, A.; Takabayashi, K.; Carson, D.A. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 1994, 368, 753–756.
- Miyakoshi, J.; Dobler, K.D.; Allalunis-Turner, J.; McKean, J.D.; Petruk, K.; Allen, P.B.; Aronyk, K.N.; Weir, B.; Huyser-Wierenga, D.; Fulton, D. Absence of IFNA and IFNB genes from human malignant glioma cell lines and lack of correlation with cellular sensitivity to interferons. Cancer Res. 1990, 50, 278–283.
- Arap, W.; Nishikawa, R.; Furnari, F.B.; Cavenee, W.K.; Huang, H.-J.S. Replacement of the pl6/CDKN2 Gene Suppresses Human Glioma Cell Growth. Cancer Res. 1995, 55, 1351–1354.
- Nishikawa, R.; Furnari, F.B.; Lin, H.; Arap, W.; Berger, M.S.; Cavenee, W.K.; Su Huang, H.J. Loss of P16INK4 expression is frequent in high grade gliomas. Cancer Res. 1995, 55, 1941–1945.
- Liban, T.J.; Thwaites, M.J.; Dick, F.A.; Rubin, S.M. Structural Conservation and E2F Binding Specificity within the Retinoblastoma Pocket Protein Family. J. Mol. Biol. 2016, 428, 3960–3971.
- Zhang, T.; Prives, C. Cyclin A-CDK Phosphorylation Regulates MDM2 Protein Interactions*. J. Biol. Chem. 2001, 276, 29702–29710.
- Giono, L.E.; Manfredi, J.J. Mdm2 Is Required for Inhibition of Cdk2 Activity by p21, Thereby Contributing to p53-Dependent Cell Cycle Arrest. Mol. Cell Biol. 2007, 27, 4166–4178.
- Zerrouqi, A.; Pyrzynska, B.; Febbraio, M.; Brat, D.J.; Van Meir, E.G. P14ARF inhibits human glioblastoma-induced angiogenesis by upregulating the expression of TIMP3. J. Clin. Investig. 2012, 122, 1283–1295.
- Yassini, P.R.; Stickler, D.L.; Bloomfield, S.M.; Wiggins, R.C.; Konat, G.W. Glioma-stimulated chemoattraction of endothelial cells and fibroblasts in vitro: A model for the study of glioma-induced angiogenesis. Metab. Brain Dis. 1994, 9, 391–399.
- Cornelius, L.A.; Nehring, L.C.; Roby, J.D.; Parks, W.C.; Welgus, H.G. Human dermal microvascular endothelial cells produce matrix metalloproteinases in response to angiogenic factors and migration. J. Investig. Dermatol. 1995, 105, 170–176.
- Aoki, K.; Nakamura, H.; Suzuki, H.; Matsuo, K.; Kataoka, K.; Shimamura, T.; Motomura, K.; Ohka, F.; Shiina, S.; Yamamoto, T.; et al. Prognostic relevance of genetic alterations in diffuse lower-grade gliomas. Neuro Oncol. 2018, 20, 66–77.
- Suzuki, H.; Aoki, K.; Chiba, K.; Sato, Y.; Shiozawa, Y.; Shiraishi, Y.; Shimamura, T.; Niida, A.; Motomura, K.; Ohka, F.; et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 2015, 47, 458–468.
- Appay, R.; Dehais, C.; Maurage, C.-A.; Alentorn, A.; Carpentier, C.; Colin, C.; Ducray, F.; Escande, F.; Idbaih, A.; Kamoun, A.; et al. CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro Oncol. 2019, 21, 1519–1528.
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016, 164, 550–563.
- Yang, R.R.; Shi, Z.-F.; Zhang, Z.-Y.; Chan, A.K.-Y.; Aibaidula, A.; Wang, W.-W.; Kwan, J.S.H.; Poon, W.S.; Chen, H.; Li, W.-C.; et al. IDH mutant lower grade (WHO Grades II/III) astrocytomas can be stratified for risk by CDKN2A, CDK4 and PDGFRA copy number alterations. Brain Pathol. 2020, 30, 541–553.
- Reis, G.F.; Pekmezci, M.; Hansen, H.M.; Rice, T.; Marshall, R.E.; Molinaro, A.M.; Phillips, J.J.; Vogel, H.; Wiencke, J.K.; Wrensch, M.R.; et al. CDKN2A Loss Is Associated with Shortened Overall Survival in Lower Grade (World Health Organization II-III) Astrocytomas. J. Neuropathol. Exp. Neurol. 2015, 74, 442–452.
- Perry, A.; Nobori, T.; Ru, N.; Anderl, K.; Borell, T.J.; Mohapatra, G.; Feuerstein, B.G.; Jenkins, R.B.; Carson, D.A. Detection of p16 gene deletions in gliomas: A comparison of fluorescence in situ hybridization (FISH) versus quantitative PCR. J. Neuropathol. Exp. Neurol. 1997, 56, 999–1008.
- Marker, D.F.; Pearce, T.M. Homozygous deletion of CDKN2A by fluorescence in situ hybridization is prognostic in grade 4, but not grade 2 or 3, IDH-mutant astrocytomas. Acta Neuropathol. Commun. 2020, 8, 169.
- Vysis CDKN2A/CEP 9 FISH Probe Kit. Available online: https://www.molecularcatalog.abbott/int/en/Vysis-CDKN2A-CEP-9-FISH-Probe-Kit (accessed on 18 January 2023).
- Satomi, K.; Ohno, M.; Matsushita, Y.; Takahashi, M.; Miyakita, Y.; Narita, Y.; Ichimura, K.; Yoshida, A. Utility of methylthioadenosine phosphorylase immunohistochemical deficiency as a surrogate for CDKN2A homozygous deletion in the assessment of adult-type infiltrating astrocytoma. Mod. Pathol. 2021, 34, 688–700.
- Hida, T.; Hamasaki, M.; Matsumoto, S.; Sato, A.; Tsujimura, T.; Kawahara, K.; Iwasaki, A.; Okamoto, T.; Oda, Y.; Honda, H.; et al. Immunohistochemical detection of MTAP and BAP1 protein loss for mesothelioma diagnosis: Comparison with 9p21 FISH and BAP1 immunohistochemistry. Lung Cancer 2017, 104, 98–105.
- Chapel, D.B.; Schulte, J.J.; Berg, K.; Churg, A.; Dacic, S.; Fitzpatrick, C.; Galateau-Salle, F.; Hiroshima, K.; Krausz, T.; Le Stang, N.; et al. MTAP immunohistochemistry is an accurate and reproducible surrogate for CDKN2A fluorescence in situ hybridization in diagnosis of malignant pleural mesothelioma. Mod. Pathol. 2020, 33, 245–254.
- Purkait, S.; Jha, P.; Sharma, M.C.; Suri, V.; Sharma, M.; Kale, S.S.; Sarkar, C. CDKN2A deletion in pediatric versus adult glioblastomas and predictive value of p16 immunohistochemistry. Neuropathology 2013, 33, 405–412.
- Schmidt, E.E.; Ichimura, K.; Reifenberger, G.; Collins, V.P. CDKN2 (p16/MTS1) gene deletion or CDK4 amplification occurs in the majority of glioblastomas. Cancer Res. 1994, 54, 6321–6324.
- Giani, C.; Finocchiaro, G. Mutation rate of the CDKN2 gene in malignant gliomas. Cancer Res. 1994, 54, 6338–6339.
- Moulton, T.; Samara, G.; Chung, W.Y.; Yuan, L.; Desai, R.; Sisti, M.; Bruce, J.; Tycko, B. MTS1/p16/CDKN2 lesions in primary glioblastoma multiforme. Am. J. Pathol. 1995, 146, 613–619.
- Sonoda, Y.; Yoshimoto, T.; Sekiya, T. Homozygous deletion of the MTS1/p16 and MTS2/p15 genes and amplification of the CDK4 gene in glioma. Oncogene 1995, 11, 2145–2149.
- Ono, Y.; Tamiya, T.; Ichikawa, T.; Kunishio, K.; Matsumoto, K.; Furuta, T.; Ohmoto, T.; Ueki, K.; Louis, D.N. Malignant astrocytomas with homozygous CDKN2/p16 gene deletions have higher Ki-67 proliferation indices. J. Neuropathol. Exp. Neurol. 1996, 55, 1026–1031.
- Rich, J.N.; Hans, C.; Jones, B.; Iversen, E.S.; McLendon, R.E.; Rasheed, B.K.A.; Dobra, A.; Dressman, H.K.; Bigner, D.D.; Nevins, J.R.; et al. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005, 65, 4051–4058.
- Zolota, V.; Tsamandas, A.C.; Aroukatos, P.; Panagiotopoulos, V.; Maraziotis, T.; Poulos, C.; Scopa, C.D. Expression of cell cycle inhibitors p21, p27, p14 and p16 in gliomas. Correlation with classic prognostic factors and patients’ outcome. Neuropathology 2008, 28, 35–42.
- Cancer Genome Atlas Research Network; Brat, D.J.; Verhaak, R.G.W.; Aldape, K.D.; Yung, W.K.A.; Salama, S.R.; Cooper, L.A.D.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498.
- Roy, D.M.; Walsh, L.A.; Desrichard, A.; Huse, J.T.; Wu, W.; Gao, J.; Bose, P.; Lee, W.; Chan, T.A. Integrated genomics for pinpointing survival loci within arm-level somatic copy number alterations. Cancer Cell 2016, 29, 737–750.
- Mohile, N.A.; Messersmith, H.; Gatson, N.T.; Hottinger, A.F.; Lassman, A.; Morton, J.; Ney, D.; Nghiemphu, P.L.; Olar, A.; Olson, J.; et al. Therapy for Diffuse Astrocytic and Oligodendroglial Tumors in Adults: ASCO-SNO Guideline. J. Clin. Oncol. 2022, 40, 403–426.
- Iwadate, Y. Epithelial-mesenchymal transition in glioblastoma progression. Oncol. Lett. 2016, 11, 1615–1620.
More
