Biofouling is the growth of organisms on wet surfaces. Biofouling includes micro- (bacteria and unicellular algae) and macrofouling (mussels, barnacles, tube worms, bryozoans, etc.) and is a major problem for industries. However, the settlement and growth of some biofouling species, like oysters and corals, can be desirable. Thus, it is important to understand the process of biofouling in detail. Modern “omic” techniques, such as metabolomics, metagenomics, transcriptomics, and proteomics, provide unique opportunities to study biofouling organisms and communities and investigate their metabolites and environmental interactions. Because "omics" originate from biomedical research and especially work at the cellular level, the learning curve for work in the environment is steep. RWesearchers envision that as use of "omics" techniques especially combining different "omics" to address complex issues like biofouling will be transformational.
- fouling
- antifouling
- metagenomic
- metabolomic
- transcriptomic
[1]1. Introduction
| Sequencing Generation | Tools Used | Features | Propose of Study | Example of Publications |
|---|---|---|---|---|
| 1st sequencing generation | Sanger sequencing | Uses capillary electrophoresis | Sequencing of genes; Identification of single biofouling organisms; Full genome sequencing |
[5][6][7][54,55,56] |
| 2nd sequencing generation | Pyrosequencing MiSeq; HiSeq; Ion Torrent |
Uses labeled nucleotides or detection of hydrogen or light | Identification of microbes in biofilms; Identification of genes | [8][9][10][11][12][39,40,41,42,43] |
| 3rd sequencing generation | Oxford Nanopore | No need for PCR amplification | Full genome sequencing; Identification of microbes in biofilms | [13][57] |
2. Metagenomics of Biofilms on Man-Made Substrata
3. Metagenomics of Biofilms on Antifouling Coatings and Biocides
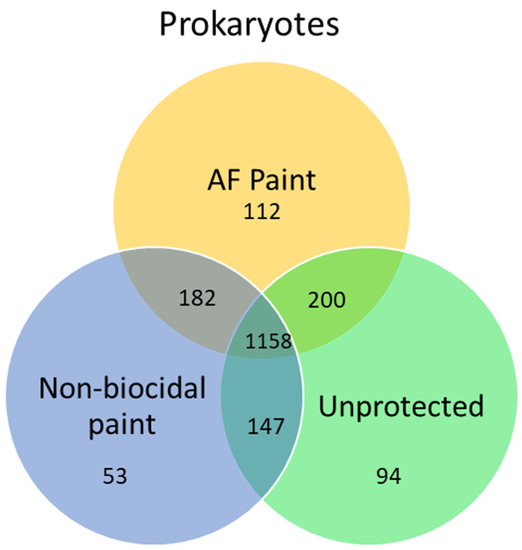
4. Environmental DNA (e-DNA)
References
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic Identification and in Situ Detection of Individual Microbial Cells without Cultivation. Microbiol. Rev. 1995, 59, 143–169. Dobretsov, Sergey, Rittschof, Daniel “Omics” Techniques Used in Marine Biofouling Studies. IJMS 2023, 24, 10518, https://doi.org/10.3390/ijms241310518.
- Escobar-Zepeda, A.; Vera-Ponce de León, A.; Sanchez-Flores, A. The Road to Metagenomics: From Microbiology to DNA Sequencing Technologies and Bioinformatics. Front. Genet. 2015, 6, 155161.
- Machida, R.J.; Knowlton, N. PCR Primers for Metazoan Nuclear 18S and 28S Ribosomal DNA Sequences. PLoS ONE 2012, 7, e46180.
- Schmidt, T.M.; DeLong, E.F.; Pace, N.R. Analysis of a Marine Picoplankton Community by 16S RRNA Gene Cloning and Sequencing. J. Bacteriol. 1991, 173, 4371–4378.
- Devereux, R.; Mundfrom, G.W. A Phylogenetic Tree of 16S RRNA Sequences from Sulfate-Reducing Bacteria in a Sandy Marine Sediment. Appl. Environ. Microbiol. 1994, 60, 3437–3439.
- Huang, Y.-L.; Li, M.; Yu, Z.; Qian, P.-Y. Correlation between Pigmentation and Larval Settlement Deterrence by Pseudoalteromonas Sp. Sf57. Biofouling 2011, 27, 287–293.
- Egan, S.; James, S.; Holmström, C.; Kjelleberg, S. Correlation between Pigmentation and Antifouling Compounds Produced by Pseudoalteromonas Tunicata. Environ. Microbiol. 2002, 4, 433–442.
- Dobretsov, S.; Abed, R.M.M.; Voolstra, C.R. The Effect of Surface Colour on the Formation of Marine Micro and Macrofouling Communities. Biofouling 2013, 29, 617–627.
- Zhang, Q.; Jie, Y.W.; Loong, W.L.C.; Zhang, J.; Fane, A.G.; Kjelleberg, S.; Rice, S.A.; McDougald, D. Characterization of Biofouling in a Lab-Scale Forward Osmosis Membrane Bioreactor (FOMBR). Water Res. 2014, 58, 141–151.
- Guo, X.; Miao, Y.; Wu, B.; Ye, L.; Yu, H.; Liu, S.; Zhang, X.-X. Correlation between Microbial Community Structure and Biofouling as Determined by Analysis of Microbial Community Dynamics. Bioresour. Technol. 2015, 197, 99–105.
- Zhang, L.; Xu, L.; Graham, N.; Yu, W. Unraveling Membrane Fouling Induced by Chlorinated Water Versus Surface Water: Biofouling Properties and Microbiological Investigation. Engineering 2022, 15, 154–164.
- Yang, J.-L.; Feng, D.-D.; Liu, J.; Xu, J.-K.; Chen, K.; Li, Y.-F.; Zhu, Y.-T.; Liang, X.; Lu, Y. Chromosome-Level Genome Assembly of the Hard-Shelled Mussel Mytilus Coruscus, a Widely Distributed Species from the Temperate Areas of East Asia. GigaScience 2021, 10, giab024.
- Colston, S.M.; Ellis, G.A.; Kim, S.; Wijesekera, H.W.; Leary, D.H.; Lin, B.; Kirkup, B.C.; Hervey, W.J.; Vora, G.J. Complete Genome Sequences of Two Bioluminescent Vibrio Campbellii Strains Isolated from Biofouling Communities in the Bay of Bengal. Genome Announc. 2018, 6, e00422-18.
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next-Generation Sequencing Technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59.
- Ma, J.; Wang, Z.; Yang, Y.; Mei, X.; Wu, Z. Correlating Microbial Community Structure and Composition with Aeration Intensity in Submerged Membrane Bioreactors by 454 High-Throughput Pyrosequencing. Water Res. 2013, 47, 859–869.
- Papadatou, M.; Robson, S.C.; Dobretsov, S.; Watts, J.E.M.; Longyear, J.; Salta, M. Marine Biofilms on Different Fouling Control Coating Types Reveal Differences in Microbial Community Composition and Abundance. Microbiologyopen 2021, 10, e1231.
- Muthukrishnan, T.; Abed, R.M.M.; Dobretsov, S.; Kidd, B.; Finnie, A.A. Long-Term Microfouling on Commercial Biocidal Fouling Control Coatings. Biofouling 2014, 30, 1155–1164.
- Winfield, M.O.; Downer, A.; Longyear, J.; Dale, M.; Barker, G.L.A. Comparative Study of Biofilm Formation on Biocidal Antifouling and Fouling-Release Coatings Using next-Generation DNA Sequencing. Biofouling 2018, 34, 464–477.
- Dobretsov, S.; Abed, R.M.M.; Muthukrishnan, T.; Sathe, P.; Al-Naamani, L.; Queste, B.Y.; Piontkovski, S. Living on the Edge: Biofilms Developing in Oscillating Environmental Conditions. Biofouling 2018, 34, 1064–1077.
- Antunes, J.T.; Sousa, A.G.G.; Azevedo, J.; Rego, A.; Leão, P.N.; Vasconcelos, V. Distinct Temporal Succession of Bacterial Communities in Early Marine Biofilms in a Portuguese Atlantic Port. Front. Microbiol. 2020, 11, 1938.
- Sushmitha, T.J.; Rajeev, M.; Murthy, P.S.; Ganesh, S.; Toleti, S.R.; Pandian, S.K. Bacterial Community Structure of Early-Stage Biofilms Is Dictated by Temporal Succession Rather than Substrate Types in the Southern Coastal Seawater of India. PLoS ONE 2021, 16, e0257961.
- Vaksmaa, A.; Egger, M.; Lüke, C.; Martins, P.D.; Rosselli, R.; Asbun, A.A.; Niemann, H. Microbial Communities on Plastic Particles in Surface Waters Differ from Subsurface Waters of the North Pacific Subtropical Gyre. Mar. Pollut. Bull. 2022, 182, 113949.
- Walter, J.M.; de Oliveira, L.S.; Tschoeke, D.A.; Meirelles, P.M.; Neves, M.H.C.B.; Batista, D.; Carvalho, A.P.; Santos Costa, R.D.; Dobretsov, S.; Coutinho, R.; et al. Metagenomic Insights Into Ecosystem Function in the Microbial Mats of a Large Hypersaline Coastal Lagoon System. Front. Mar. Sci. 2021, 8, 715335.
- Lema, K.A.; Constancias, F.; Rice, S.A.; Hadfield, M.G. High Bacterial Diversity in Nearshore and Oceanic Biofilms and Their Influence on Larval Settlement by Hydroides Elegans (Polychaeta). Environ. Microbiol. 2019, 21, 3472–3488.
- Guo, Z.; Wang, L.; Cong, W.; Jiang, Z.; Liang, Z. Comparative Analysis of the Ecological Succession of Microbial Communities on Two Artificial Reef Materials. Microorganisms 2021, 9, 120.
- Ribani, A.; Schiavo, G.; Utzeri, V.J.; Bertolini, F.; Geraci, C.; Bovo, S.; Fontanesi, L. Application of next Generation Semiconductor Based Sequencing for Species Identification and Analysis of Within-Species Mitotypes Useful for Authentication of Meat Derived Products. Food Control 2018, 91, 58–67.
- Harb, M.; Xiong, Y.; Guest, J.; Amy, G.; Hong, P.-Y. Differences in Microbial Communities and Performance between Suspended and Attached Growth Anaerobic Membrane Bioreactors Treating Synthetic Municipal Wastewater. Environ. Sci. Water Res. Technol. 2015, 1, 800–813.
- Liu, Y.; Jeraldo, P.; Mendes-Soares, H.; Masters, T.; Asangba, A.E.; Nelson, H.; Patel, R.; Chia, N.; Walther-Antonio, M. Amplification of Femtograms of Bacterial DNA Within 3 h Using a Digital Microfluidics Platform for MinION Sequencing. ACS Omega 2021, 6, 25642–25651.
- Colston, S.M.; Hervey, W.J.; Horne, W.C.; Haygood, M.G.; Petersen, B.D.; van Kessel, J.C.; Vora, G.J. Complete Genome Sequence of Vibrio Campbellii DS40M4. Microbiol. Resour. Announc. 2019, 8, e01187-18.
- Hosoe, A.; Suenaga, T.; Sugi, T.; Iizumi, T.; Nagai, N.; Terada, A. Complete Genome Sequence of Pseudomonas Putida Strain TS312, Harboring an HdtS-Type N-Acyl-Homoserine Lactone Synthase, Isolated from a Paper Mill. Microbiol. Resour. Announc. 2020, 9, e00055-20.
- Li, Y.-F.; Chen, Y.-R.; Yang, J.-L.; Bao, W.-Y.; Guo, X.-P.; Liang, X.; Shi, Z.-Y.; Li, J.-L.; Ding, D.-W. Effects of Substratum Type on Bacterial Community Structure in Biofilms in Relation to Settlement of Plantigrades of the Mussel Mytilus Coruscus. Int. Biodeterior. Biodegrad. 2014, 96, 41–49.
- Petersen, L.-E.; Moeller, M.; Versluis, D.; Nietzer, S.; Kellermann, M.Y.; Schupp, P.J. Mono- and Multispecies Biofilms from a Crustose Coralline Alga Induce Settlement in the Scleractinian Coral Leptastrea Purpurea. Coral Reefs 2021, 40, 381–394.
- Cacabelos, E.; Ramalhosa, P.; Canning-Clode, J.; Troncoso, J.S.; Olabarria, C.; Delgado, C.; Dobretsov, S.; Gestoso, I. The Role of Biofilms Developed under Different Anthropogenic Pressure on Recruitment of Macro-Invertebrates. Int. J. Mol. Sci. 2020, 21, 2030.
- Dey, S.; Rout, A.K.; Behera, B.K.; Ghosh, K. Plastisphere Community Assemblage of Aquatic Environment: Plastic-Microbe Interaction, Role in Degradation and Characterization Technologies. Environ. Microbiome 2022, 17, 32.
- Zhurina, M.V.; Bogdanov, K.I.; Gannesen, A.V.; Mart’yanov, S.V.; Plakunov, V.K. Microplastics as a New Ecological Niche For Multispecies Microbial Biofilms within the Plastisphere. Microbiology 2022, 91, 107–123.
- Singh, S.P.; Sharma, P.; Bano, A.; Nadda, A.K.; Varjani, S. Microbial Communities in Plastisphere and Free-Living Microbes for Microplastic Degradation: A Comprehensive Review. Green Anal. Chem. 2022, 3, 100030.
- Wright, R.J.; Erni-Cassola, G.; Zadjelovic, V.; Latva, M.; Christie-Oleza, J.A. Marine Plastic Debris: A New Surface for Microbial Colonization. Environ. Sci. Technol. 2020, 54, 11657–11672.
- Briand, J.-F.; Barani, A.; Garnier, C.; Réhel, K.; Urvois, F.; LePoupon, C.; Bouchez, A.; Debroas, D.; Bressy, C. Spatio-Temporal Variations of Marine Biofilm Communities Colonizing Artificial Substrata Including Antifouling Coatings in Contrasted French Coastal Environments. Microb. Ecol. 2017, 74, 585–598.
- Sathe, P.; Laxman, K.; Myint, M.T.Z.; Dobretsov, S.; Richter, J.; Dutta, J. Bioinspired Nanocoatings for Biofouling Prevention by Photocatalytic Redox Reactions. Sci. Rep. 2017, 7, 3624.
- von Ammon, U.; Wood, S.A.; Laroche, O.; Zaiko, A.; Tait, L.; Lavery, S.; Inglis, G.; Pochon, X. The Impact of Artificial Surfaces on Marine Bacterial and Eukaryotic Biofouling Assemblages: A High-Throughput Sequencing Analysis. Mar. Environ. Res. 2018, 133, 57–66.
- Beata, G. The Use of -Omics Tools for Assessing Biodeterioration of Cultural Heritage: A Review. J. Cult. Herit. 2020, 45, 351–361.
- Ward, C.S.; Diana, Z.; Ke, K.M.; Orihuela, B.; Schultz, T.P.; Rittschof, D. Microbiome Development of Seawater-Incubated Pre-Production Plastic Pellets Reveals Distinct and Predictive Community Compositions. Front. Mar. Sci. 2022, 8, 807327.
- Joardar, I.; Dutta, S. A Selective Review on the Novel Approaches and Potential Control Agents of Anti-Biofouling and Anti-Biofilming. Appl. Biochem. Biotechnol. 2022.
- Xia, Z.; Gu, J.; Wen, Y.; Cao, X.; Gao, Y.; Li, S.; Haffner, G.D.; MacIsaac, H.J.; Zhan, A. EDNA-Based Detection Reveals Invasion Risks of a Biofouling Bivalve in the World’s Largest Water Diversion Project. Ecol. Appl. 2023, e2826.
- Ibabe, A.; Rayón, F.; Martinez, J.L.; Garcia-Vazquez, E. Environmental DNA from Plastic and Textile Marine Litter Detects Exotic and Nuisance Species Nearby Ports. PLoS ONE 2020, 15, e0228811.
- Bowers, H.A.; Pochon, X.; von Ammon, U.; Gemmell, N.; Stanton, J.-A.L.; Jeunen, G.-J.; Sherman, C.D.H.; Zaiko, A. Towards the Optimization of EDNA/ERNA Sampling Technologies for Marine Biosecurity Surveillance. Water 2021, 13, 1113.
- Pawlowski, J.; Bruce, K.; Panksep, K.; Aguirre, F.I.; Amalfitano, S.; Apothéloz-Perret-Gentil, L.; Baussant, T.; Bouchez, A.; Carugati, L.; Cermakova, K.; et al. Environmental DNA Metabarcoding for Benthic Monitoring: A Review of Sediment Sampling and DNA Extraction Methods. Sci. Total Environ. 2022, 818, 151783.
