1. Loncg Non-Coding RNAs in Adult Gliomas
Adult gliomas primarily affect individuals over the age of 18, with the majority occurring in older adults. The most common adult glioma is glioblastoma multiforme, which is an aggressive and malignant tumor
[38][1]. The CNS5 classification encompasses several types of adult gliomas with varying levels of malignancy. Each glioma type in the CNS5 classification has distinct histopathological features and grading criteria, which take into account factors such as nuclear atypia, mitotic activity, microvascular proliferation, and the presence of necrosis
[10][2]. In addition to histological characteristics, the WHO classification also emphasizes the importance of molecular markers for certain glioma types, such as IDH mutation status, 1p/19q codeletion, and MGMT promoter methylation, which can have prognostic and predictive implications. Non-coding RNAs (ncRNAs) have been reported to play significant roles in glioma progression by regulating gene expression, cellular processes, and the tumor microenvironment
[26][3]. LncRNAs can regulate gene expression through diverse mechanisms, including acting as scaffolds for protein complexes, interacting with DNA, RNA, or proteins, and modulating chromatin structure or transcriptional regulation
[39][4]. Several lncRNAs have been implicated in glioma growth and invasion. Some lncRNAs promote glioma cell proliferation, survival, migration, and invasion by interacting with specific signaling pathways, such as the Wnt/β-catenin pathway or the PI3K/AKT pathway
[40,41][5][6]. LncRNAs were also found to participate in epigenetic modifications, such as DNA methylation and histone modifications, influencing gene expression patterns in gliomas
[42][7].
Different regulation mechanisms exerted by lncRNAs have been reported to influence adult glioma angiogenesis. LncRNAs can act as competing endogenous RNAs (ceRNAs) by sequestering miRNAs, thereby preventing their binding to target mRNAs. This interaction can modulate the expression of target genes and affect glioma progression
[43][8]. Various lncRNAs have been identified as oncogenes or tumor suppressors in adult gliomas. For example, MALAT1, HOTAIR, and H19 have been found to promote glioma cell proliferation, migration, invasion, and angiogenesis, thereby acting as oncogenes
[44,45,46][9][10][11]. Conversely, lncRNAs such as MEG3 and GAS5 have shown tumor-suppressive effects by inhibiting glioma cell growth and inducing apoptosis in adults
[47][12]. Some lncRNAs can bind to proteins and modulate their activity, including transcription factors involved in angiogenesis-related gene expression. By recruiting transcription factors or interacting with other proteins, lncRNAs can regulate pathways that contribute to tumor angiogenesis. LncRNAs can also interact with other RNA molecules, such as mRNAs or miRNAs, to influence their stability, translation, or activity. This interaction can indirectly affect angiogenesis-related processes and pathways. Furthermore, lncRNAs can regulate the behavior and function of neighboring cells, including those within the tumor microenvironment. By influencing the communication between tumor cells and surrounding cells, lncRNAs can impact angiogenesis by promoting or inhibiting the formation of new blood vessels
[48,49,50][13][14][15]. LncRNAs have essential roles in adult glioma cell proliferation and migration and guide gene regulation during tumor angiogenesis. They can directly regulate the cytoskeleton or influence the cytoskeleton through various signaling pathways during tumor migration
[51][16]. Tang et al.
[52][17] revealed that the long non-coding RNA-activated by transforming growth factor β (
ATB) promoted the invasion of adults glioma cells through nuclear factor kappa B (NF-κB) and p38 mitogen-activated protein kinase (MAPK) pathways. Other lncRNAs were found to sponge miRNAs
[53][18]. Dysregulated lncRNAs in gliomas have been associated with clinicopathological features, patient survival, and therapeutic response. They hold potential as diagnostic biomarkers and therapeutic targets for glioma treatment in adults. LncRNAs can modulate gene expression through epigenetic mechanisms in adult glioma. They can interact with chromatin-modifying proteins, such as polycomb repressive complexes (PRC1 and PRC2), to regulate the methylation and acetylation status of histones, thereby impacting gene transcription
[54][19]. LncRNA HOTAIR is known to interact with PRC2 to silence tumor suppressor genes and promote adult glioma progression
[55][20]. LncRNAs can regulate key signaling pathways involved in adult glioma pathogenesis, such as the PI3K/AKT, Wnt/β-catenin, and Notch pathways. They can modulate the expression or activity of pathway components, affecting downstream signaling events and promoting glioma cell survival, proliferation, and invasion. LncRNA CCAT2 has been found to activate the Wnt/β-catenin pathway in glioma cells, contributing to tumor growth
[56][21]. While
ou
r understanding of the precise functions and mechanisms of lncRNAs in adult glioma is still evolving, the emerging evidence suggests their important roles in glioma pathogenesis and their potential as therapeutic targets and biomarkers. Further research is needed to unravel the intricate interactions and regulatory networks involving lncRNAs in adult gliomas, paving the way for novel therapeutic strategies and improved patient outcomes.
2. Loncg Non-Coding RNAs in Pediatric Gliomas
Malignant brain and central nervous system (CNS) tumors account for ~21% of tumors in children and represent the second leading cause of pediatric cancer deaths after leukemia
[57,58][22][23]. Pediatric glioma, a common central nervous system tumor in children, is classified into low-grade and high-grade categories. Low-grade gliomas refer to benign, slow-growing grade I or II lesions, while high-grade gliomas encompass rapidly progressing tumors such as anaplastic astrocytoma (grade III), glioblastoma, and diffuse intrinsic pontine glioma (grade IV)
[59][24]. These non-coding RNA molecules have been found to be dysregulated in pediatric gliomas and contribute to tumor development and progression. Various lncRNAs have been identified as tumor suppressors or oncogenes in pediatric gliomas. For example, LINC00312 and MEG3 have been found to act as tumor suppressors, inhibiting glioma cell proliferation and inducing apoptosis
[60][25]. On the other hand, lncRNAs such as MALAT1 and HOTAIR have been implicated as oncogenes, promoting glioma cell growth, invasion, and angiogenesis
[61][26]. LncRNAs can modulate gene expression through epigenetic mechanisms in pediatric gliomas. They can interact with chromatin-modifying proteins and affect the methylation and acetylation status of histones, thereby regulating gene transcription. LncRNA H19, for instance, has been shown to play a role in the epigenetic silencing of tumor suppressor genes in pediatric gliomas
[62][27]. LncRNAs can function as ceRNAs (competing endogenous RNAs) by sponging miRNAs and preventing them from binding to their target mRNAs. This interaction can impact the expression of target genes and affect glioma cell behavior. LncRNA HULC, for example, has been found to regulate miRNA-mediated gene expression in pediatric gliomas
[63][28]. LncRNAs participate in the regulation of signaling pathways that are crucial for pediatric glioma pathogenesis. They can interact with key components of pathways, such as the PI3K/AKT and Wnt/β-catenin pathways, influencing downstream signaling events and cellular processes. LncRNA CCAT2 has been shown to activate the Wnt/β-catenin pathway in pediatric gliomas, promoting tumor growth
[64][29]. Dysregulated expression of specific lncRNAs in pediatric gliomas has been associated with tumor grade, prognosis, and treatment response. These lncRNAs also hold promise as diagnostic and prognostic biomarkers for pediatric gliomas, aiding in patient stratification and personalized treatment decisions
[65][30]. Understanding the roles of lncRNAs in pediatric gliomas provides insights into the molecular mechanisms underlying tumor development and may pave the way for the development of novel therapeutic strategies and targeted therapies.
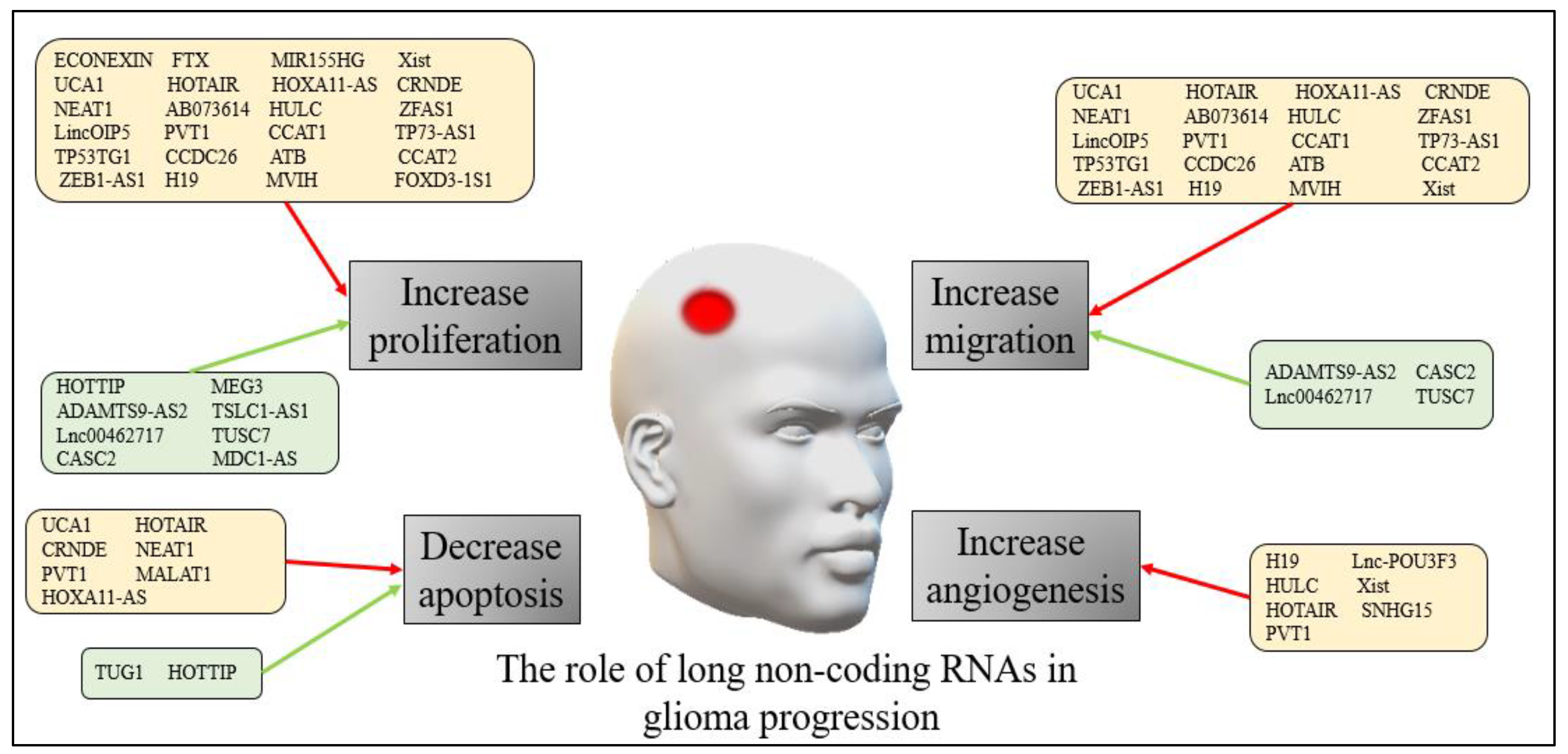
The precise mechanisms through which lncRNAs exert their effects in both adults and pediatric glioma progression are still being explored. However, accumulating evidence suggests that lncRNAs play critical roles in regulating various cellular processes involved in tumor growth, invasion, metastasis, and treatment response. Figure 1 presents a summary of the role of long non-coding RNAs in human glioma progression.
Figure 1. The role of long non-coding RNAs in human glioma progression. The red arrows refer to down-regulation of the non-coding RNAs, while the green arrows refer to up-regulation.
In high-grade gliomas, the high expression of taurine-upregulated gene 1 (
TUG1) lncRNA has been reported to favor angiogenesis by inhibiting the function of miR-299, which has a binding site for vascular endothelial growth factor (VEGF)
[66][31]. Ye et al.
[67][32] showed that the down-regulation of long intergenic non-protein coding RNA (
LINC01116) suppresses the angiogenesis of adult glioma in vivo. The same authors suggested that
LINC01116 may play a role in the post-transcriptional regulation of VEGF by binding to miR-31-5p. In the same manner, X-inactive specific transcript (
XIST) sponges miR-485 in human glioma micro-vascular endothelial cells; hence, the binding of miR-485 enables SRY-box transcription factor 7 (SOX7) to increase the expression level of VEGF
[68][33]. Additionally, Cheng et al.
[69][34] identified a correlation between lncRNA
XIST and miR-429 in adult glioma cells, which was found to highly affect angiogenesis. The authors also reported that
XIST knockdown reduced glioma angiogenesis in vitro and in vivo. Likewise, small nucleolar RNA host gene 15 (
SNHG15) lncRNA knockdown reduced the expression levels of pro-angiogenic factor cell division control protein 42 homolog (CDC42) and VEGF because the miR-153 was longer sponged and led to suppression of tumor angiogenesis in adult glioma cells
[70][35]. RNA binding proteins were found to play critical roles in almost all gene expression steps, especially in transcription as well as post-transcriptional regulations
[71][36]. Yang et al.
[72][37] investigated the expression and the role of lncRNA
LINC00346 in adults glioma angiogenesis regulation and demonstrated that ankyrin repeat and KH domain containing one protein (ANKHD1) and
LINC00346 were highly up-regulated in adult glioma cells, whereas the zinc finger protein 655 (ZNF655) expression was decreased. The authors reported that the inhibition of ANKHD1,
LINC00346, or the overexpression of ZNF655 impeded angiogenesis of the cells, suggesting that ANKHD1 targeted the long RNA
LINC00346 and enhanced its stability, which then bound to ZNF655 mRNA via their Alu elements. Thereby,
LINC00346 facilitated ZNF655 mRNA degradation via a staufen1 (STAU1) mediated mRNA decay mechanism. Furthermore, the promoter region of ANKHD1 was targeted by ZNF655 and formed the feedback loop of ANKHD1/
LINC00346/ZNF655 that regulated the angiogenesis of adult glioma cells. Refer to
Table 1 for the summary of different lncRNAs mechanisms in adult glioma progression and angiogenesis.
3. Loncg Non-Coding RNAs as Biomarkers in Adults and Pediatric Glioma Diagnosis
Long non-coding RNAs (lncRNAs) have shown promise as biomarkers for glioma diagnosis. Their dysregulated expression patterns in gliomas can serve as indicators of disease presence, classification, and prognosis
[86][51]. Dysregulated expression of specific lncRNAs has been observed in gliomas compared to normal brain tissue. This aberrant expression can be detected using various techniques, such as quantitative polymerase chain reaction (qPCR), microarray analysis, or RNA sequencing
[87][52]. The expression levels of certain lncRNAs have demonstrated diagnostic potential for differentiating gliomas from non-neoplastic brain tissue or other brain tumor types. Poor outcome of high-grade glioma patients has been linked to late-stage diagnosis and treatment due to
our low knowledge about the early stage of glioma, which cannot be observed with conventional diagnostic approaches
[88][53]. However, even in aggressive therapy of human glioma with a combination of different treatment modalities many types of gliomas have been reported with having a pessimistic prognosis for survival
[89,90][54][55]. The current gold standard for categorizing nervous system tumors, as outlined in the 2021 edition of the World Health Organization (WHO) classification, prioritizes molecular diagnosis over histologic diagnosis. In many countries, histological evaluation is still in use for different glioma diagnoses, which is difficult and sometimes not clear to acquire tissue because of its special anatomical position in the CNS. Furthermore, diagnosis of glioma using imaging methods such as magnetic resonance imaging and computed tomography before clinical diagnosis or treatment are widely used, but they have failed to improve the early diagnosis rate, which results in significant glioma spreading
[91][56]. Recently, some tumor-related molecules, including different non-coding RNAs, have been reported to be significantly involved in the angiogenesis and progression of gliomas in pediatrics and have been investigated for being diagnostic or prognostic biomarkers
[92][57]. LncRNAs have shown the ability to distinguish between different glioma subtypes, aiding in their accurate classification. For example, lncRNA expression profiles have been used to differentiate between glioblastoma and lower-grade gliomas (LGG), as well as to identify specific molecular subtypes within GBM
[93][58]. The sensitivity, accuracy, and specificity of these molecular biomarkers are highly adequate for the evaluation of early diagnosis or prognosis of human glioma with reliable clinical significance. lncRNAs have multiple biological functions, including transcriptional control, chromatin remodeling, and post-transcriptional processing
[94,95][59][60]. The expression levels of certain lncRNAs have been associated with the prognosis and overall survival of glioma patients. High expression of some lncRNAs has been correlated with poorer outcomes, including shorter survival times and increased tumor aggressiveness
[96][61]. These lncRNAs can serve as prognostic biomarkers, providing valuable information for patient risk stratification and treatment decision-making. LncRNAs have the potential to predict the response of gliomas to specific treatments. For example, the expression levels of certain lncRNAs have been associated with resistance to chemotherapy or radiotherapy. Monitoring the expression of these lncRNAs could help identify patients who are more likely to benefit from certain treatment strategies or require alternative therapeutic approaches. Another advantage of lncRNAs as biomarkers is their potential for non-invasive detection. lncRNAs can be detected in various body fluids, including blood, cerebrospinal fluid (CSF), and urine
[98][62]. This enables the development of minimally invasive or liquid biopsy-based approaches for glioma diagnosis and monitoring, reducing the need for invasive procedures, such as surgical biopsies.
Expression abnormalities of lncRNAs and miRNAs have been reported to be associated with the gliomas progression in both adults and pediatrics
[100][63]. Recent studies have indicated that
MALAT1 lncRNA has promising potential to serve as a diagnostic biomarker for gliomas and other cancers with sufficient specificity and sensitivity
[26,101][3][64]. Up-regulation of the
HOXA-AS3 lncRNA significantly promoted tumor progression, and it can be used to predict poor prognosis in glioma
[102][65]. Wang et al.
[103][66] investigated the role of the
LINC00152 in high-grade glioma in adults and observed its expression was independently associated with poor prognosis of high-grade glioma. The authors also reported that the overall survival of the high-expression glioma group was significantly shorter than the low-expression group, while the knockdown of
LINC00152 inhibited tumor growth in vivo, suggesting
LINC00152 as a potential prognostic biomarker for high-grade glioma, especially in adults. Recent investigations have demonstrated the role of antisense lncRNAs in numerous biological processes, including tumor cell proliferation, invasion, survival, migration, and even apoptosis through multiple epigenetic modifications and chromatin remodeling
[104,105][67][68]. It was reported that HAND2 antisense RNA 1 (
HAND2-AS1) in endometrioid endometrial carcinoma was significantly down-regulated, and its overexpression considerably inhibited invasion and metastasis of tumor through inactivating neuromedin U
[106][69]. In a different study, Han et al.
[107][70] found that the
HOXA11-AS lncRNA could serve as a diagnostic target and biomarker for glioma in adults as well as pediatrics. Similarly, Wu et al.
[108][71] used RNA sequencing data to profile the differentially expressed antisense lncRNAs in various gliomas gathered from the
Chinese Glioma Genome Atlas database and found convincing evidence of the possibility of using WDFY3 antisense RNA 2 (
WDFY3-AS2) lncRNA as a novel valuable prognostic biomarker and diagnostic agent for diffuse glioma in adults. Wang et al.
[109][72] analyzed expression profiles of four lncRNA, namely, AGAP2 antisense RNA 1 (
AGAP2-AS1), long intergenic non-protein coding RNA 1198 (
LINC01198), TPT1 antisense RNA 1 (
TPT1-AS1) and MIR155 host gene (
MIR155HG) of anaplastic astrocytomas patients. The authors found that
TPT1-AS1 was postulated to have a significant protective role, while the other three (
AGAP2-AS1,
LINC01198, and
MIR155HG) were suggested to be involved in tumor development as they were expressed more in the high-risk group.
4. Long LncNon-Coding RNAs in Adults and Pediatric Glioma Therapy
Long non-coding RNAs (lncRNAs) have emerged as potential therapeutic targets in glioma therapy. Their dysregulation in gliomas and involvement in various cellular processes make them attractive candidates for novel treatment strategies. LncRNAs affect cell behavior in several different ways, from epigenetic regulation, including recruitment of transcription factor, splicing regulation, and messenger RNA stability and translation
[110][73]. Owing to the ability of lncRNAs to interact with DNA, RNA, and proteins, lncRNAs also involve chromatin modification, post-transcriptional modifications, and scaffolding
[111][74]. Chen et al.
[112][75] found that up-regulation of the expression of carbamoyl phosphate synthetase 1 intronic transcript 1 (
CPS1-IT1) lncRNA resulted in a significant decrease in glioma proliferation, migration, and invasion abilities in adults, suggesting
CPS1-IT1 as anti-oncogenic molecules in glioma treatment. In a different study,
lncGRS-1 was found to be a glioma-specific therapeutic target in both adults and pediatrics. Using antisense oligonucleotides that targeted
lncGRS-1, it was able to selectively decrease tumor growth and sensitize glioma cells to radiotherapy
[113][76]. Ji et al.
[114][77] found that the knockdown of the SWI/SNF complex antagonist associated with prostate cancer 1 (
SChLAP1) lncRNA suppressed the growth of glioma in adults as it was found to be highly expressed. The authors reported that
SChLAP1 was stabilized by its binding to heterogeneous nuclear ribonucleoprotein L (HNRNPL), which significantly enhanced the interaction with the protein alpha-actinin-4 (ACTN4). Urothelial carcinoma-associated 1 (UCA1) lncRNA was recently discovered as a proto-oncogene in the progression of many cancers, including glioma in adults and pediatrics
[115][78]. UCA1 could promote glioma cell proliferation and cell cycle in adults through the up-regulation of cyclin D1 transcription. Targeting dysregulated lncRNAs through their silencing or inhibition is a potential therapeutic approach. This can be achieved using antisense oligonucleotides (ASOs), small interfering RNAs (siRNAs), short hairpin RNAs (shRNAs), or CRISPR/Cas9-mediated gene editing
[116,117,118][79][80][81]. By reducing the expression or function of oncogenic lncRNAs, these strategies aim to inhibit glioma growth, invasion, and other malignant characteristics. Some lncRNAs act as key regulators of gene expression in gliomas. Targeting these lncRNAs can modulate the expression of downstream genes involved in glioma progression. By manipulating the expression or activity of specific lncRNAs, it is possible to influence critical pathways associated with tumor growth, angiogenesis, invasion, and therapy resistance. Huang et al.
[119][82] up-regulated UCA1 in adult glioma cell lines and tissues using lentiviral vector transfections and observed a significant elevation in glioma cell invasion through the induction of epithelial-mesenchymal transition. The same authors reported that the growth of adult glioma cells was significantly suppressed upon the silencing of UCA1 in vivo, and the expression of circadian locomotor output cycles kaput (CLOCK) was also significantly lowered after UCA1 knockdown, suggesting the utility of targeting this lncRNA for adult glioma treatment.
Certain lncRNAs have been reported to participate in epigenetic regulation, influencing chromatin structure and gene expression patterns in gliomas
[120][83]. Targeting these lncRNAs can alter the epigenetic landscape and restore normal gene expression profiles. This approach holds promise for reversing aberrant gene silencing and promoting tumor-suppressive pathways in glioma cells. Combining lncRNA-targeted therapies with other treatment modalities, such as chemotherapy or radiotherapy, is being explored. By targeting lncRNAs alongside traditional treatments, it may be possible to enhance treatment efficacy, overcome therapy resistance, and improve patient outcomes.
LncRNA H19 has been reported in numerous publications as well as its role in promoting cancer cell growth and proliferation
[121,122][84][85] by regulating the proteins that are involved in apoptosis, cell cycle regulation as well as glucose metabolism, and chemoresistance
[123][86].
H19 is expressed in both hepatocellular carcinoma and adult glioma-associated endothelial cells within the exosomes, which promote tumor angiogenesis
[124][87]. Zhao et al.
[125][88] observed that the overexpression of
H19 significantly promoted glioma cell proliferation and migration in adults. The autophagy of the tested cell lines was enhanced after the silencing of
H19 and suppressed when the lncRNA was overexpressed.
H19 overexpression inhibited the mechanistic target of rapamycin (mTOR) protein phosphorylation and promoted Unc-51-like kinase 1 (ULK1) phosphorylation. The regulation of mTOR signaling was found to be the mechanism by which
H19 promotes the proliferation, migration, and autophagy of adult glioma cells
[125][88]. In a different study, Sheng et al.
[126][89] recently reported that p53 targeted lncRNA suppressor of tumorigenicity 7 antisense RNA 1 (
ST7-AS1) acted as a significant tumor suppressor by interacting with polypyrimidine tract-binding protein 1 (PTBP1) to suppress the Wnt/β-catenin signaling pathway in adult and pediatric glioma. They contributed to significant inhibition of glioma cells migration, invasion, and proliferation, in addition to promoting the apoptosis of glioma cells. Down-regulation of PTBP1 occurred as a result of
ST7-AS1 direct binding at the post-transcriptional level. Certain lncRNAs have been implicated in conferring resistance to therapies in gliomas
[127][90]. Targeting these lncRNAs can sensitize glioma cells to standard treatments, making them more responsive to chemotherapy or radiation. This approach aims to enhance the effectiveness of existing therapies and improve treatment responses. In addition to targeting lncRNAs, certain lncRNAs themselves can be used as therapeutic agents. For example, engineered lncRNAs can be designed to exert specific functions, such as acting as decoys for miRNAs or as competitors for protein-protein interactions. These engineered lncRNAs can potentially modulate critical pathways and disrupt glioma progression.
Table 2 provides a summary of studies that involved the manipulation of lncRNAs activity for glioma therapy.