Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Sirius Huang and Version 1 by Gyanendra Kumar Rai.
Among all the abiotic factors, drought is likely to have one of the most detrimental effects on soil organisms and plants. Drought is a major problem for crops because it limits the availability of water, and consequently nutrients which are crucial for plant growth and survival. This results in reduced crop yields, stunted growth, and even plant death, according to the severity and duration of the drought, the plant’s developmental stage, and the plant’s genetic background. The ability to withstand drought is a highly complex characteristic that is controlled by multiple genes, making it one of the most challenging attributes to study, classify, and improve.
- abiotic stress
- drought tolerance
- agriculture
- yield
- osmotic stress
1. Detrimental Consequences of Drought Conditions on Plant
Abiotic stresses generated by various climatic conditions can have a detrimental impact on crop development and production. Plants adapt to numerous abiotic challenges by undergoing morphological, physio-biochemical, as well as cellular changes [9][1]. Drought is one of the major constraints for crop output worldwide because it negatively affects crop efficiency as well as compromising its productivity [10][2]. Water shortage triggers a variety of crop reactions at physio-biochemical, molecular, as well as morphological planes, inevitably distressing crop yield [11][3] by impacting different functions, as illustrated in Figure 1. Even a short dry spell negatively impacts plant water dynamics during plant growth, which then disrupts the whole metabolic activity, both at the molecular and physiological levels, depending on the degree and extent of drought [12,13][4][5]. When there is a water shortage, one of the primary drivers of the response of plants at the cellular level is indirect or direct oxidative stress. This results in the modification of metabolic machinery as well as the destabilization of membrane stability, which causes extreme metabolic concerns and drastically alters plant activity [14,15][6][7]. Drought is acknowledged as a constraint in many aspects of crop production. One of the most basic factors of crop development is germination, which influences overall plant health, and is highly vulnerable to drought. Substantial variations in germination rates have indeed been confirmed for numerous crops, viz., wheat, sorghum, and maize. Further, under the influence of water stress, plant growth is often reduced with or without any sign of leaf wilting in the initial phases of vegetative growth, and flowering can be disrupted [16][8]. Due to low soil moisture during drought, crops frequently have trouble in absorbing nutrients, which inhibits stem growth; reduction in shoot length was evident under water shortages in the studies [17][9]. Drought is a limiting factor that impacts several physiological processes in plants, especially metabolism, and proliferation. On the other hand, plant drought tolerance responses are activated simultaneously, encompassing a variety of biological mechanisms that are active at different stages of plant growth and operate somewhere at the level of cells, tissues, and, ultimately, the entire plant. Specifically, plants increase water transport and absorption by growing a much more productive, deep, and wide root system while they prevent water loss by maintaining an optimum rate of transpiration [13][5]. Additionally, the growth of drought-resilient crops is aided by the employment of crop growth regulators, membrane coherence retention, ideal plant cultivars, antioxidants, proteins related to drought stress, and ion channel proteins such as aquaporins [18][10].
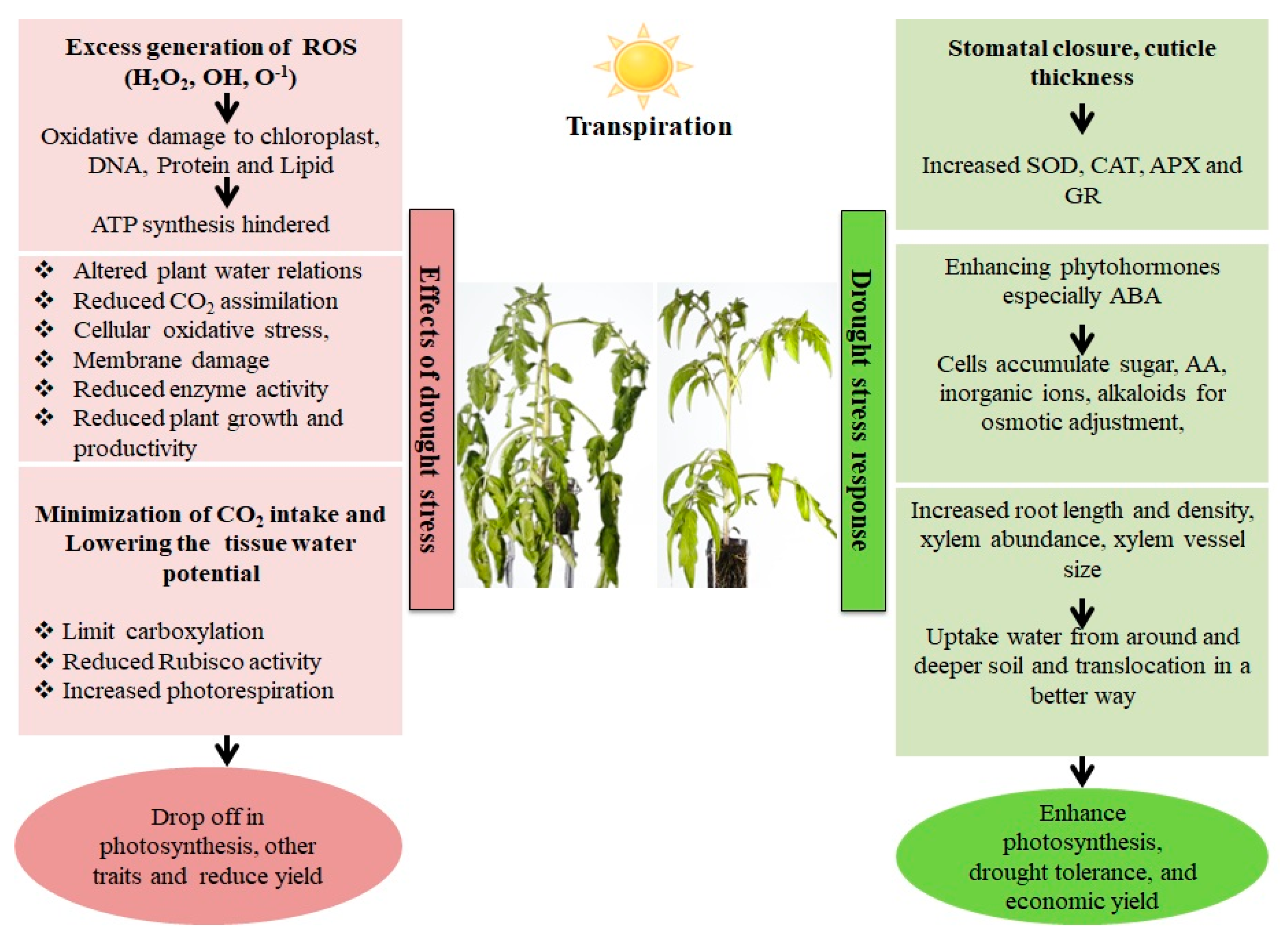
Figure 1. Effect and response of a plant in a drought stress environment. Drought stress causes an imbalance between electron excitation and utilization during photosynthesis, resulting in the generation of reactive oxygen species (ROS), predominantly superoxide (O2−) and hydrogen peroxide. (H2O2). These ROS cause oxidative stress by damaging cell membranes, proteins, and nucleic acids. Plants have both enzymatic as well as non-enzymatic detoxification mechanisms to scavenge ROS viz., SOD (Superoxide dismutase), which catalyzes the conversion of O2− into the least reactive H2O2. The H2O2 is detoxified into O2 and H2O via the enzymatic activities of Catalase (CAT) and Ascorbate Peroxidase (APX). Non-enzymatic antioxidants involved in cellular defense include carotenoids and glutathione (GSH). Carotenoids defend the photosynthetic machinery byphotoprotection, ROS scavenging, membrane stabilization and contributing to the regeneration of other antioxidants, while GSH protects the chloroplasts from ROS damage bydetoxification, and protection against lipid peroxidation.
2. Molecular Aspects Underlying Resilience of Crops against Drought
Enhancing agronomic attributes of crops that would provide resistance to multiple abiotic and biotic stresses has always indeed been a worldwide concern [19][11]. The understanding of global warming and climate change emphasizes the importance of incorporating specific, realistic, and sustainable strategies. Crop yield sustainability amid drought seems to be an important concern in some countries. Drought intensity varies over time and space, so to endure stress; plants have adapted complicated mechanisms with diverse physiological and morphological approaches [20][12]. Crops confront drought through different degrees of adaptation, avoidance, and evasion [21][13]. Exploiting genetic characteristics which boost drought tolerance while maintaining high yield is critical during plant management. Drought resistance of wheat, soybean, rice, and maize might be improved using recombinant and classic breeding approaches. Previously, traditional breeding remained the most fruitful method of growing plants, promoting their growth in water-stressed ecosystems. Those very same strategies, however, seem to be labor and time intensive, as well as costly. Under environmental stresses, molecular markers have already played a vital role in attempting to portray plant genetic variability [22][14]. Several quantitative trait loci (QTLs) associated with boosting drought resilience have been already identified in various crops. However, the precision and reliability of QTL recognition continue to be an issue [23][15]. Considering this, genome editing seems to have been extremely successful in enhancing crop resistance to abiotic and biotic stresses [24][16]. Technological advances that can broaden the response of crops to stress as well as make them more environmentally friendly are needed. The advent of genome editing techniques has already resulted in major breakthroughs in plant breeding. Genetic manipulation tools employ sequence-specific nucleases to incorporate recognized variations into the genome [25][17]. CRISPR-Cas gene-editing systems have attracted widespread praise for their versatility and easiness of use. The whole strategic approach utilizes a guide RNA and an intricate endonuclease (Cas endonuclease) that alters DNA strands to generate double-stranded DNA breaks. Such breaks are then restored via endogenic cellular repair mechanisms, resulting in the generation of novel genetic variations [26][18]. Competently, the CRISPR-Cas platform has been employed to achieve tolerance against a multitude of abiotic stresses, which include toxicity of heavy metal ions, salinity, drought, and submergence [27][19]. The ongoing review provides an overview of the use of the CRISPR/Cas9 platform in plants to accomplish drought resistance, as well as explores the technology’s potential towards the increment of drought-tolerant plant varieties. Comprehensive molecular research findings have elucidated to decipher the cellular mechanisms that control plant drought response. Abscisic acid (ABA) modulates plant drought response by limiting stomatal conductance and gene expression to restrict water loss via transpiration [12][4]. The transcription factor basic leucine zipper (bZIP), also known as ABA-responsive element (ABRE)-binding proteins, is required for ABA signaling [28][20]. AREB1 increased expression (ABF2) enhanced drought resistance in soybean, rice, and Arabidopsis, whereas AREB1 failure promoted drought vulnerability [29][21]. Moreover, during drought, AREB1 regulates a broad array of genes downstream of the ABA signal transduction pathway, including ABA-mediated antioxidant signaling, ABA biogenesis, and osmotic stress response. As a result, AREB1 is indeed an interesting candidate for improving the responses of plants to drought [30][22]. The availability of genome sequences for many plants, as well as breakthroughs in gene editing techniques, had already opened new opportunities for breeding for a variety of attractive characteristics. Technological advancements in gene editing, such as transcription activator-like effector nucleases (TALENs) and zinc-finger nucleases (ZFNs), have also enabled molecular biologists to target any desired gene even further precisely [31][23]. Nonetheless, the above strategies are costly as well as time-consuming since because they involve complex stages encompassing protein designing. Unlike first-generation gene-editing initiatives, CRISPR/Cas9 method includes simple cloning methods and convenient implementation. This very same Cas9 is being experimented with several different guide RNAs to target multiple sites in the genomic DNA. Regarding concrete evidence experiments in plants with a preparatory CRISPR/Cas9 unit, a variety of Cas9 endonucleases (StCas9, SaCas9, and NmCas9) have already been introduced to optimize target precision and decrease off-target cleavage. Moreover, the accessibility of Cas9 enzymes from some of the other bacteria has improved gene editing effectiveness and accuracy [32][24]. The current review summarizes the crop augmentation options that are available to agro-biotechnologists using well-established Cas9-based gene-editing strategies. Cas9 enzymes have been utilized to improve resistance to biotic and abiotic stresses. Incorporating these techniques is anticipated to result in non-genetically altered (non-GMO) crops with the desired phenotype, which might also increase yield under biotic and abiotic stress conditions [21][13].
3. CRISPR/Cas9-Based Precise Genome Editing for Crop Drought Resilience
Multiple molecular investigations have shown that the ABA is the primary component of plant drought responses, controlling both the expression of target genes relating to stress and stomatal conductance, which prevents water loss. The essential elements of ABA signaling are indeed the binding domain (ABRE) and bZIP unit (AREBs/ABFs) of transcriptional regulators, also known as ABA-responsive factors [33][25]. Whereas the AREB1 inactivation increased susceptibility to drought, AREB1 overexpression increased their ability to withstand it. AREB1 is a crucial component of the osmotic stress response, antioxidant signaling, and ABA biogenesis [34][26]. It controls a diverse range of gene expression across the ABA signal transduction pathway. As a direct consequence, AREB1 might be regarded as a crucial target for boosting plant drought resilience (Figure 2 and Figure 3). To unlock the promoter region of AREB1 in Arabidopsis, a customized CRISPR-Cas9 system combining sgRNA, its catalytic subunit of the HAT enzyme, and dead Cas9 (dCas9) was adopted [35][27]. Apparently, acetylation of the core histone that resulted from the engagement of the Arabidopsis HAT catalytic site increased the AREB1 promoter area’s responsiveness to the transcriptional zone. Mutants had greater levels of AREB1 transcription, stomatal conductance, as well as chlorophyll despite drought, according to physiological and molecular studies. Furthermore, in the presence of water stress, AREB1 induced RD29A transcription [35][27]. Under drought stress, the recombinant CRISPR lines showed enhanced survival rates.
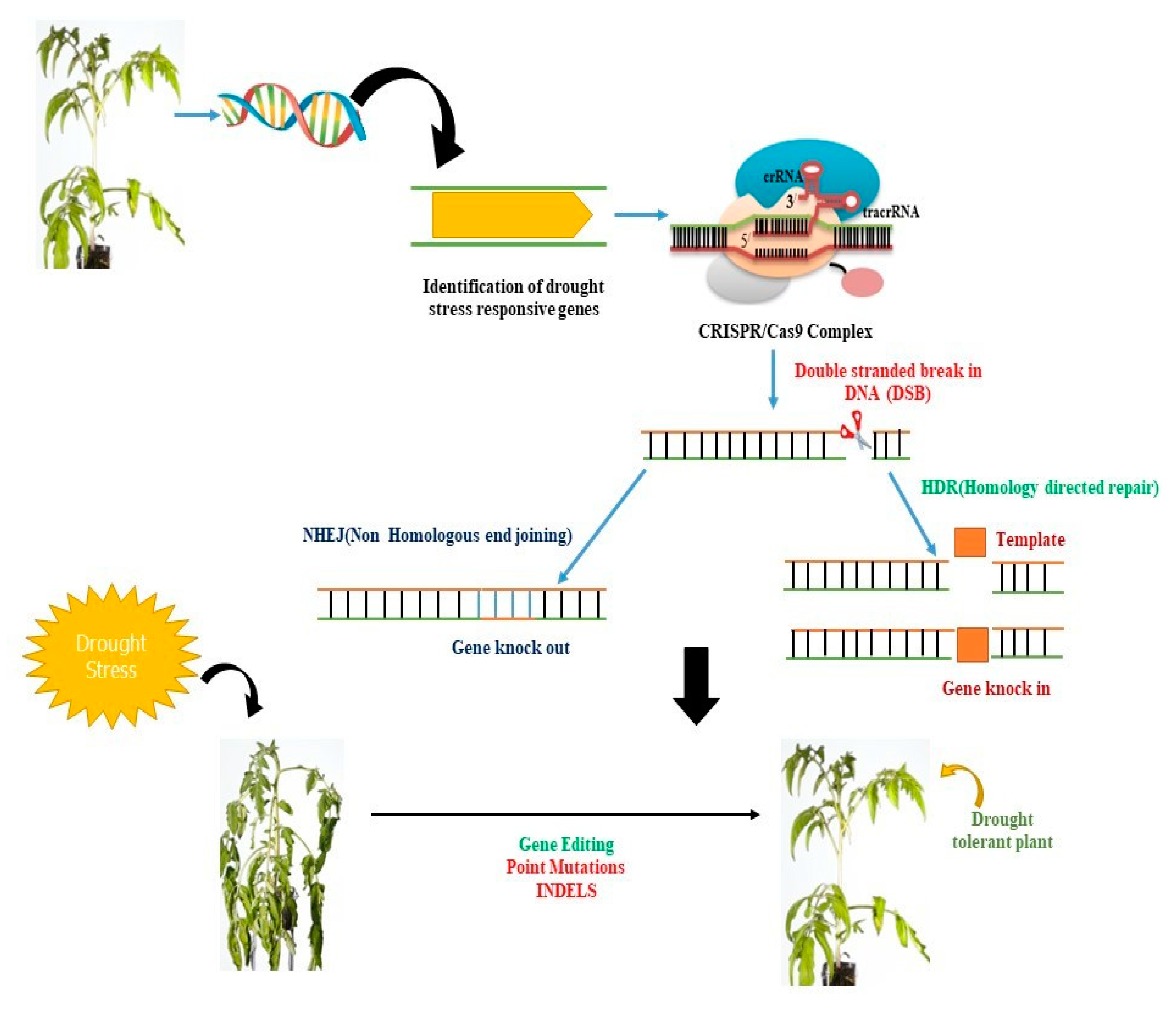
Figure 2. CRISPR/Cas9-mediated genetic manipulation can be used to enhance plant productivity under stress conditions. The Cas9 protein can be guided by a single guide RNA (sgRNA) to a specific genomic region of interest. The CRISPR/Cas9 system then identifies a G-rich protospacer adjacent motif (PAM) region at the proximal end of the target DNA and cleaves it, creating a blunt-ended double-stranded break (DSB). These DSBs can be repaired by the plant’s endogenous repair system via non-homologous end joining (NHEJ) or homology-directed repair. CRISPR/Cas9 can induce mutations through insertions or deletions (INDELs), gene deletions, or multiplex gene knockout, providing a powerful tool for genetic manipulation in plants.
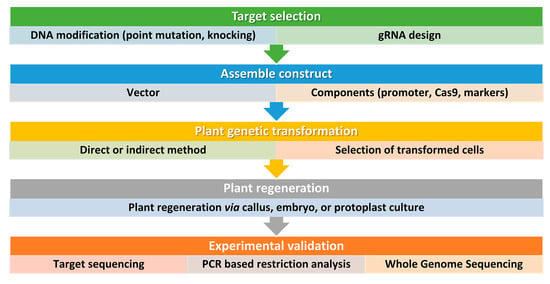
Figure 3. Simplified workflow for CRISPR/Cas9 genome editing in plants. This workflow outlines the key steps involved in using this technology for plant biotechnology. It is important to consider all factors before starting and to design and implement screening procedures also beyond the DNA analysis of the transformed plants. Step 1: Selection of target sequence. The first step in CRISPR/Cas9 genome editing is to select the target sequence. The aim is typically to generate point mutations or small insertions/deletions that result in gene knockout or loss of function. An accurate guide RNA (gRNA) design is carried out to maximize efficiency and minimize the risk of off-target mutations. Step 2: Vector design and assembly. Before constructing a vector, several factors should be considered, including the techniques used for plant genetic transformation and the aim of the study. Several vectors are available and can be tailored for a specific application. For instance, Cas9 and gRNA can be generated from the same vector or separate vectors, and, the Cas9 and gRNA expression can be driven by different promoters according to the plant species and aims. The most used proteins are based on the type IIA Cas9 from Streptococcus pyogenes. The native Cas9 coding sequence has been codon optimized for monocots or dicots. Step 3: DNA delivery. Delivering DNA into plant cells is performed using conventional methods in plant biotechnology, such as Agrobacterium-mediated transformation, biolistic microparticle bombardment, or protoplast transformation, followed by plant regeneration when necessary (Step 4). Step 5: Screening. The screening of plant DNA follows standard procedures and may include whole genome sequencing to check for off-target mutations, especially if back-crossing is not a viable option.
Overall, these findings show that drought-responsive genes may be positively controlled by the CRISPR-Cas system to effectively elicit epigenetic modifications for improving plant drought tolerance. The primary activator of the ABA-dependent high osmotic stress response and signaling is SNF1 associated protein kinase 2 (SnRK2), a class of protein kinases peculiar to plants [36][28]. Seed germination, response to hyperosmotic stress, ABA-mediated stomatal closure, ABA signaling, drought resistance, and plant growth are all processes that are affected by the frequent involvement of SnRK2 members. The activation of drought-regulated genes by AtSnRK2.8 in Arabidopsis really demonstrated a characteristic stress regulatory network to positively influence drought resistance [37][29]. AREB-ABF axis and its receptors were controlled by SnRK2 irrespective of the fact the absence of appreciable changes in stomatal damage as well as mortality seen between the wild-type and the SnRK2.8 mutant, according to microarray analysis. Similarly, plant growth improvements and abiotic stress response were shown in rice when SnRK2 family members of sub-class I and III were present [38][30].
References
- Ansari, W.A.; Atri, N.; Singh, B.; Pandey, S. Changes in antioxidant enzyme activities and gene expression in two muskmelon genotypes under progressive water stress. Biologia Plantarum. 2017, 61, 333–341.
- Rai, G.K.; Parveen, A.; Jamwal, G.; Basu, U.; Kumar, R.R.; Rai, P.K.; Sharma, J.P.; Alalawy, A.I.; Al-Duais, M.A.; Hossain, M.A.; et al. Leaf Proteome Response to Drought Stress and Antioxidant Potential in Tomato (Solanum lycopersicum L.). Atmosphere 2021, 12, 1021.
- Zlatev, Z.; Lidon, F.C. An overview on drought induced changes in plant growth, water relations and photosynthesis. Emirates J. Food Agric. 2012, 57–72.
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.S.P. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86.
- Ansari, W.A.; Atri, N.; Singh, B.; Kumar, P.; Pandey, S. Morpho-physiological and biochemical responses of muskmelon genotypes to different degree of water deficit. Photosynthetica 2018, 56, 1019–1030.
- Sharma, A.; Zheng, B. Melatonin mediated regulation of drought stress: Physiological and molecular aspects. Plants 2019, 8, 190.
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, oxidative damage and oxygen deprivation stress: A review. Ann. Bot. 2003, 91, 179–194.
- Bhatt, R.M.; Rao, N.S. Influence of pod load on response of okra to water stress. Indian J. Plant Physiol. 2005, 10, 54.
- Razmjoo, K.; Heydarizadeh, P.; Sabzalian, M.R. Effect of salinity and drought stresses on growth parameters and essential oil content of Matricaria chamomile. Int. J. Agric. Biol. 2008, 10, 451–454.
- Hoang, T.M.L.; Tran, T.N.; Nguyen, T.K.T.; Williams, B.; Wurm, P.; Bellairs, S.; Mundree, S. Improvement of salinity stress tolerance in rice: Challenges and opportunities. Agronomy 2016, 6, 54.
- Ansari, W.A.; Chandanshive, S.U.; Bhatt, V.; Nadaf, A.B.; Vats, S.; Katara, J.L.; Sonah, H.; Deshmukh, R. Genome editing in cereals: Approaches, applications and challenges. Int. J. Mol. Sci. 2020, 21, 4040.
- Ansari, W.A.; Atri, N.; Pandey, M.; Singh, A.K.; Singh, B.; Pandey, S. Influence of drought stress on morphological, physiological and biochemical attributes of plants: A review. Biosci. Biotechnol. Res. Asia. 2019, 16, 697–709.
- Sourour, A.; Afef, O.; Mounir, R.; Mongi, B.Y. A review: Morphological, physiological, biochemical and molecular plant responses to water deficit stress. Int. J. Eng. Sci. 2017, 6, 1–4.
- Queiroz, M.S.; Oliveira, C.E.; Steiner, F.; Zuffo, A.M.; Zoz, T.; Vendruscolo, E.P.; Silva, M.V.; Mello, B.F.F.R.; Cabra, R.C.; Menis, F.T. Drought stresses on seed germination and early growth of maize and sorghum. J. Agric. Sci. 2019, 11, 310–318.
- Khan, A.; Pan, X.; Najeeb, U.; Tan, D.K.Y.; Fahad, S.; Zahoor, R.; Luo, H. Coping with drought: Stress and adaptive mechanisms, and management through cultural and molecular alternatives in cotton as vital constituents for plant stress resilience and fitness. Biol. Res. 2018, 51.
- Shinwari, Z.K.; Jan, S.A.; Nakashima, K.; Yamaguchi-Shinozaki, K. Genetic engineering approaches to understanding drought tolerance in plants. Plant Biotechnol. Rep. 2020, 14, 151–162.
- Haque, E.; Taniguchi, H.; Hassan, M.M.; Bhowmik, P.; Karim, M.R.; Śmiech, M.; Zhao, K.; Rahman, M.; Islam, T. Application of CRISPR/Cas9 genome editing technology for the improvement of crops cultivated in tropical climates: Recent progress, prospects, and challenges. Front. Plant Sci. 2018, 9, 617.
- Brokowski, C.; Adli, M. CRISPR ethics: Moral considerations for applications of a powerful tool. J. Mol. Biol. 2019, 431, 88–101.
- Raza, A.; Charagh, S.; Razzaq, A.; Javed, R.; Khan RS, A.; Hasanuzzaman, M. Brassicaceae plants response and tolerance to drought stress: Physiological and molecular interventions. In The Plant Family Brassicaceae: Biology and Physiological Responses to Environmental Stresses; Springer: Berlin/Heidelberg, Germany, 2020; pp. 229–261.
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170.
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. The Plant J. 2010, 61, 672–685.
- Li, X.Y.; Liu, X.; Yao, Y.; Li, Y.H.; Liu, S.; He, C.Y.; Li, J.M.; Lin, Y.Y.; Li, L. Overexpression of Arachis hypogaea AREB1 gene enhances drought tolerance by modulating ROS scavenging and maintaining endogenous ABA content. Int. J. Mol. Sci. 2013, 14, 12827–12842.
- Ansari, W.A.; Atri, N.; Yang, L.; Singh, B.; Pandey, S. Genetic diversity in muskmelon based on SSR markers and morphological traits under well-watered and water-deficit condition. Biocat. Agric. Biotechnol. 2020, 26, 101630.
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nature Biotechnol. 2014, 31, 686–688.
- Bhat, J.A.; Ali, S.; Salgotra, R.K.; Mir, Z.A.; Dutta, S.; Jadon, V.; Tyagi, A.; Mushtaq, M.; Jain, N.; Singh, P.K.; et al. Genomic selection in the era of next generation sequencing for complex traits in plant breeding. Front. Genet. 2016, 7, 221.
- Bouzroud, S.; Gasparini, K.; Hu, G.; Barbosa, M.A.M.; Rosa, B.L.; Fahr, M.; Bendaou, N.; Bouzayen, M.; Zsögön, A.; Smouni, A.; et al. Down regulation and loss of auxin response factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. Genes 2020, 11, 272.
- Paixão, J.F.; Gillet, F.X.; Ribeiro, T.P.; Bournaud, C.; Lourenço-Tessutti, I.T.; Noriega, D.D.; Melo, B.P.D.; de Almeida-Engler, J.; Grossi-de-Sa, M.F. Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a Histone AcetylTransferase. Sci. Rep. 2019, 9, 1–9.
- Bota, J.; Medrano, H.; Flexas, J. Is photosynthesis limited by decreased Rubisco activity and RuBP content under progressive water stress. New Phytol. 2004, 162, 671–681.
- Umezawa, T.; Yoshida, R.; Maruyama, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc. Nat. Acad. Sci. USA 2004, 101, 17306–17311.
- Kulik, A.; Wawer, I.; Krzywińska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 protein kinases-key regulators of plant response to abiotic stresses. Omics A J. Integr. Biol. 2011, 15, 859–872.
More
