Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Conner Chen and Version 1 by Dolores Fernández-Ortuño.
Powdery mildew and rust fungi are major agricultural problems affecting many economically important crops and causing significant yield losses. These fungi are obligate biotrophic parasites that are completely dependent on their hosts for growth and reproduction.
- RNA interference
- VIGS
- HIGS
1. Introduction
RNAi is a biological mechanism in which short noncoding RNAs (sRNAs) are used to deliberately downregulate gene expression at the transcriptional or posttranscriptional level. Posttranscriptional gene silencing is a tightly controlled system that relies on a group of proteins to coordinate gene silencing based on sequence complementarity between sRNA and target mRNA [1,2][1][2]. MicroRNAs (miRNAs) and short-interfering RNAs (siRNAs) are two types of regulatory sRNAs encoded by plants. miRNAs are 20–22 nucleotide (nt) sequences formed from a single-stranded RNA molecule that folds back on itself, creating a double-stranded region with a loop called RNA hairpin (hpRNAs), whereas siRNAs are 20–24 nt sequences derived from lengthy dsRNA precursors [3,4][3][4]. RNAi regulates a variety of biological processes, including plant immunity [5], and siRNAs and miRNAs have been identified as key factors in plant defense against viruses, bacteria, and fungi [6,7,8,9][6][7][8][9]. As shown in Figure 1, the silencing process starts with the binding of a host’s ribonuclease-III called Dicer (DICER) to long dsRNAs or hpRNA and their cleavage into siRNAs of 21–25 nt in length [10,11][10][11]. DICER has a helicase domain, a Piwi/Argonaute/Zwille (PAZ) motif, a dsRNA binding domain at the N-terminus, and two RNase III motifs at the C-terminus. DICER-generated siRNAs are subsequently integrated into the RNA-induced silencing complex (RISC). This multicomponent protein complex contains an Argonaute protein (AGO) with an sRNA-binding domain and endo-nucleolytic activity for RNA cleavage, which is triggered by the ATP-dependent unwinding of the siRNA duplex [12] (Figure 1). The passenger strand is degraded, and the guide strand binds to the target mRNA sequence and stimulates endonucleolytic cleavage or inhibits translation once the siRNA is integrated into RISC [13,14][13][14]. The existence of an RNA-dependent RNA polymerase (RdRP), which can interact with the RISC complex and create new dsRNA based on the partially degraded target template utilizing the hybridized siRNA strands as primers, is assumed to be the cause of this effect (Figure 1). Then, the DICER enzyme acts on the synthetized dsRNA to make additional siRNAs (secondary siRNAs). Once a dsRNA has been delivered into a cell, its influence can last throughout development; moreover, dsRNAs can be exported to neighboring cells, spreading the knockout gene effect throughout the organism [15]. There is growing evidence that sRNAs are mobilized in bidirectional interactions between plants and their pathogens, laying the groundwork for cross-kingdom RNAi (ck-RNAi) as a plant defensive mechanism [6,9,16][6][9][16].
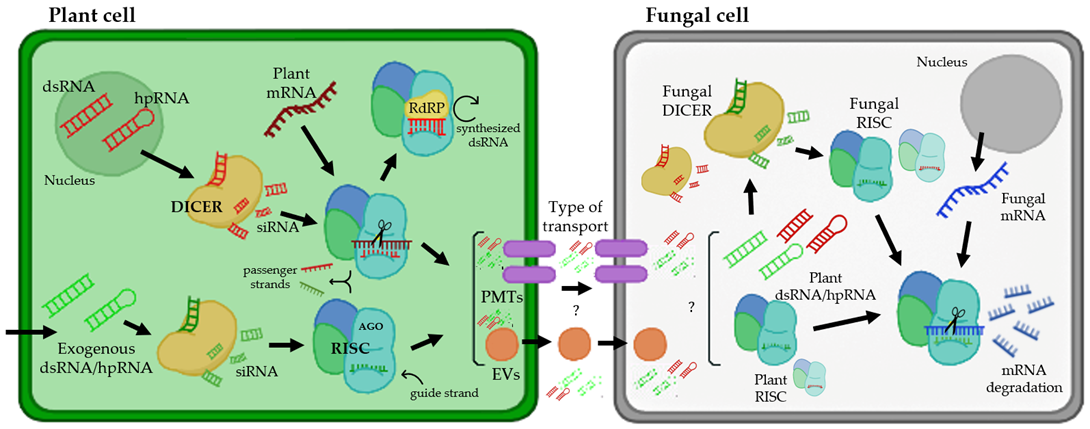
Figure 1. Interaction between a plant cell and fungal pathogen from the perspective of plant RNAi-mediated host-induced gene silencing. The arrows represent the flow of the gene silencing mechanism using RNAi. In the nucleus of a plant cell, dsRNA and hpRNA are produced as normal defense responses or from hairpin RNAs in transgenic RNAi plants (targeting a fungal gene). In addition, there are several biotechnology tools that allow the entry of exogenous dsRNAs or hpRNAs. These molecules of dsRNA and hpRNA can be processed by the DICER enzyme, creating siRNAs, which are integrated into the RNA-induced silencing complex (RISC), which contains an Argonaute protein (AGO), using them as templates for mRNA silencing. For the amplification of this silencing mechanism, there is an RNA-dependent RNA polymerase (RdRP) that can synthesize new dsRNAs using hybridized siRNA strands as primers. siRNAs produced in plant cells can be transported presumably by two types of transport (represented with question marks): via plant-derived extracellular vesicles (EVs) and plasma membrane-located transporters (PMTs). Inside the fungal cell, the mechanism of silencing works similarly to plant cells, producing the assembly of siRNAs with the RISC and inducing the silencing of specific fungal mRNAs.
Several studies have been conducted to investigate siRNA uptake in various organisms, and two primary mechanisms for host-derived RNA absorption have been proposed. One mechanism is siRNA absorption via plant-derived extracellular vesicles (EVs), which is based on the occurrence of exosome-like vesicles in plants that can carry bioactive compounds such as sRNAs to animal cells [17,18,19][17][18][19] (Figure 1). For example, in the fungal pathogen Sclerotinia sclerotiorum, using live cell images, it was concluded that the uptake of dsRNA occurs via clathrin-mediated endocytosis [20]. The other proposed mechanism occurs via plasma membrane-located transporters (Figure 1). This mechanism was supported by a study with the transmembrane protein SID-1, expressed in Drosophila S2 cells, which enabled passive dsRNA uptake from a culture medium [21]. Later, the lysosome transmembrane protein SIDT2 was identified in mammals and was shown to be involved in RNA uptake and subsequent degradation in this organelle [22].
This process of RNA trafficking from plant host cells to interacting pathogens has also been described in a variety of plant pathogenic fungi and oomycetes, such as Botrytis cinerea, Cochliobolus sativus, Fusarium graminearum, Plasmopara viticola, Podosphaera xanthii, Sclerotinia sclerotiorum and Venturia inaequalis [23,24,25,26,27,28,29,30][23][24][25][26][27][28][29][30]. Currently, the mechanisms of the transfer of sRNAs from plants to pathogenic fungi are unknown; however, the discovery that these eukaryotic pathogens are inhibited by sRNAs targeting their essential and/or pathogenicity genes has raised the possibility that plants could be protected by a new generation of environmentally friendly RNA-based fungicides that can be extremely specific and easily adaptable to control multiple diseases at the same time [31].
2. Powdery Mildew and Rust Fungi
Obligate biotrophic fungi are a group of the most damaging plant pathogens, incurring massive economic losses and jeopardizing global food security. Powdery mildew and rust fungi infect more than 10,000 plant species, including many agronomically important crops, such as cereals, grapevines, many vegetables, and fruits, as well as ornamental and forest plants [32]. Their complete dependence on the host to feed, grow, and reproduce significantly complicates their manipulation under laboratory conditions, hindering research on their lifestyle and pathogenicity mechanisms at the molecular level [33].
Powdery mildew fungi are phytopathogenic ascomycetes belonging to the Erysiphaceae family, order Erysiphales, which includes 900 species and more than 80 genera. They cause damage in a wide range of angiosperm hosts, including both monocotyledons and dicotyledons plants. The fungal pathogens belonging to this group are easily identified by their symptoms, including the presence of powdery white patches on leaf surfaces, petioles, stems, blooms, and even fruits [34,35][34][35] (Figure 2A). In general, powdery mildew fungi exhibit both asexual and sexual life cycles (Figure 2B). The latter is highly uncommon for some species and only occurs under suitable environmental and nutritional conditions [36]. The asexual cycle starts after a conidium settles on a susceptible host plant. After its germination, it forms a small primary germ tube that elongates to become an appressorium (Figure 2B), which is in charge of penetrating the cuticle [37]. Subsequently, a hyphal peg will penetrate the epidermal cell creating a primary haustorium [38]. Upon effective infection, the main hyphae will branch and generate secondary hyphae and secondary haustoria. Later, conidiophores will emerge vertically from hyphae, generating a varying number of conidia or asexual spores depending on the species [36,39,40,41,42][36][39][40][41][42] (Figure 2B). This epiphytic fungal growth causes typical powdery mildew disease signs. In the event of sexual reproduction, two mating opposite hyphae need to be in contact to create a fruiting body termed chasmothecium, which holds one or more ascus containing the ascospores or sexual spores (Figure 2B) [41,42][41][42]. Although the exact infection structures developed by ascospores have not yet been determined, it is assumed that they are similar to those developed by conidia [35,43][35][43].
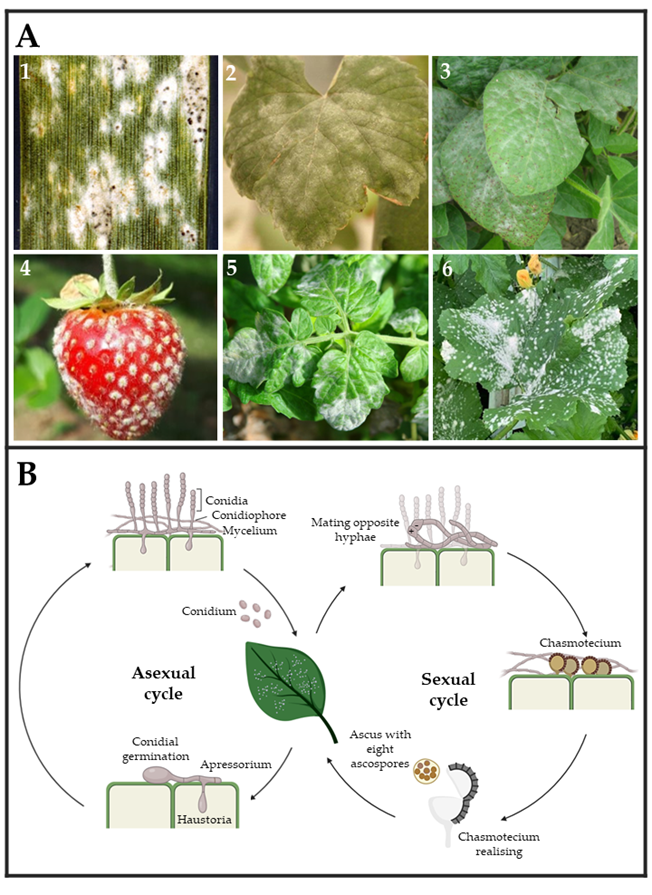
Figure 2. (A) Powdery mildew symptoms observed on leaves and fruits of several crops. (1) Wheat (Triticum aestivum) leaf, (2) wine grape (Vitis vinifera) leaf, (3) soybean (Glycine max) leaves, (4) strawberry (Fragaria sp.), (5) tomato (Solanum lycopersicum) leaves, and (6) melon (Cucumis melo) leaves infected by Blumeria graminis, Erysiphe necator, Microsphaera diffusa, Podosphaera aphanis, Leveillula taurica, and Podosphaera xanthii, respectively. Pictures (1)–(6) were taken by Clemson University—USDA CES, Yuan-Min Shen (National Taiwan University), Daren Mueller (Homemade, Bugwood.org (accessed on 28 March 2023)), University of Hertfordshire, Scot Nelson (Homemade flickr.com (accessed on 28 March 2023), respectively. (B) The typical powdery mildew life cycle is divided into two types of reproduction. Asexual reproduction is carried out by the release of conidium spores, which develop haustoria capable of acquiring nutrients from plant cells and giving rise to hyphae and conidiophores. Sexual reproduction occurs when two hyphae from opposing mating types form a chasmothecium capable of releasing an ascus with eight ascospores.
On the other hand, rust fungi comprise two orders, Uredinales and Pucciniales, in the widely varied phylum of Basidiomycota formed by mushrooms and bracket fungi. Rust fungi are divided into 14 families and 166 genera. Most species are found in the genera Puccinia and Uromyces, which have approximately 5000 and 1500 taxon names listed in Index Fungorum 2013, respectively [44]. Like powdery mildews, rusts are obligate biotrophic and pathogenic fungi that live on vascular plants ranging from ferns to monocots and gymnosperms to angiosperms (Figure 3A) [45,46,47,48][45][46][47][48]. Rust fungi have a typical macrocyclic-heteroecious life cycle where meiosis occurs in short-lived basidia formed by germinating teliospores (Figure 3B). Haploid basidiospores infect the aecial host and develop protoaecia and pycnia, among other fungal structures (Figure 3(B1,B2)). Pycnial nectar droplets create haploid pycniospores and receptive hyphae, where fertilization can take place between spores, and receptive hyphae of suitable mating types (Figure 3(B2,B3)). Following plasmogamy, dikaryotic aecia differentiate inside the host, and aeciospores are liberated and distributed by the wind (Figure 3(B3)). Aeciospores infect the telial host, causing the production of uredinia and urediniospores, which is followed by recurrent cycles of vegetative development on the telial host for several weeks or months, usually throughout the summer. Uredinia develops into telia in early fall, going through an overwintering phase during which karyogamy occurs, resulting in diploid dormant teliospores Figure 3(B4,B5) [45,48,49][45][48][49].
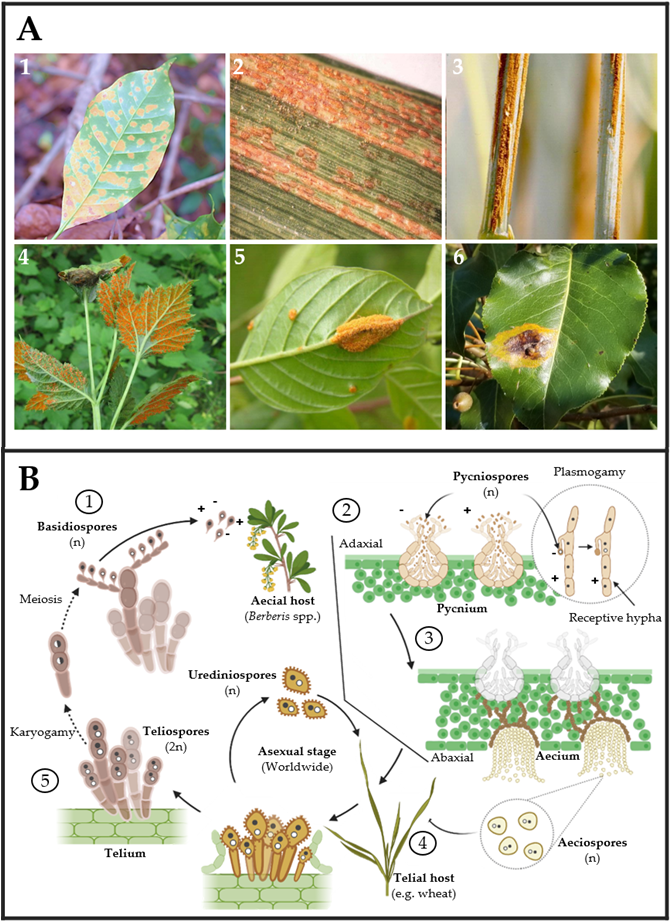
Figure 3. (A) Rust fungi symptoms observed on leaves of several crops: (1) coffee leaf (Coffea arabica), (2) barley leaf (Hordeum vulgare), (3) oat stem (Avena sativa), (4) black raspberry leaves (Rubus occidentalis), (5) glossy buckthorn (Frangula alnus), and (6) pear leaves (Pyrus spp.) infected by Hemileia vastatrix, Puccinia striiformis, P. graminis f. sp. avenae, Arthuriomyces peckianus, Puccinia coronata and Gymnosporangium sabinae, respectively. Pictures (1)–(6) were taken by Dr. Parthasarathy Seethapathy (Amrita School of Agricultural Sciences), Mary Burrows (Montana State University), Howard F. Schwartz (Colorado State University), Sandra Jensen (Cornell University), Milan Zubrik (Forest Research Institute—Slovakia; homemade, Bugwood.org (accessed on 28 March 2023)) and Sue Muller (Homemade MarylandBiodiversityProject.com (accessed on 28 March 2023)), respectively. (B) The typical rust fungal life cycle includes two types of hosts. First, aecial hosts are infected by haploid basidiospores ①, which generate pycnium as reproductive structures ②. Pycnium produce pycniospores with different polarities, which produce plasmogamy for generating haploid aeciospores ③. Second, aeciospores infect the telial host, in which urediniospores can be generated for asexual reproduction ④. In the telial host, teliospores are produced by karyogamy. Finally, basidiospores are produced by the meiosis of teliospores ⑤.
Both powdery mildew and rust fungi share a special structure of parasitism developed inside plant cells termed the haustorium. This specialized cell has been shown to deploy effectors, which are secreted proteins translocated into the plant cell, responsible for promoting the manipulation of the plant’s immune system and orchestrating the reprogramming of gene expression from the infected tissue to maintain fungal growth and development upon a successful infection [50,51][50][51]. The haustorium is also involved in the uptake of nutrients such as carbohydrates and amino acids and potentially water from the host via ion pumps present in the plasma membrane [52]. In addition, its ability to take genetic material such as dsRNA or siRNA makes it a key element in the development of methods of genetic transformation for biotrophic fungi, opening a world of possibilities that will allow many processes and functions to be studied in depth in the future [53,54,55][53][54][55].
References
- Gheysen, G.; Vanholme, B. RNAi from plants to nematodes. Trends Biotechnol. 2007, 25, 89–92.
- Shabalina, S.A.; Koonin, E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008, 23, 578–587.
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741.
- D’Ario, M.; Griffiths-Jones, S.; Kim, M. Small RNAs: Big impact on plant development. Trends Plant Sci. 2017, 22, 1056–1068.
- Deng, Y.; Wang, J.; Tung, J.; Liu, D.; Zhou, Y.; He, S.; Du, Y.; Baker, B.; Li, F. A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 2018, 14, e1006756.
- Hua, C.; Zhao, J.H.; Guo, H.S. Trans-kingdom RNA silencing in plant–fungal pathogen interactions. Mol. Plant 2018, 11, 235–244.
- Rosa, C.; Kuo, Y.W.; Wuriyanghan, H.; Falk, B.W. RNA interference mechanisms and applications in plant pathology. Annu. Rev. Phytopathol. 2018, 56, 581–610.
- Muhammad, T.; Zhang, F.; Zhang, Y.; Liang, Y. RNA interference: A natural immune system of plants to counteract biotic stressors. Cells 2019, 8, 38.
- Schaefer, L.K.; Parlange, F.; Buchmann, G.; Jung, E.; Wehrli, A.; Herren, G.; Müller, M.C.; Stehlin, J.; Schmid, R.; Wicker, T.; et al. Cross-kingdom RNAi of pathogen effectors leads to quantitative adult plant resistance in wheat. Front. Plant Sci. 2020, 11, 253.
- Zrachya, A.; Kumar, P.P.; Ramakrishnan, U.; Levy, Y.; Loyter, A.; Arazi, T.; Lapidot, M.; Gafni, Y. Production of siRNA targeted against TYLCV coat protein transcripts leads to silencing of its expression and resistance to the virus. Transgenic Res. 2007, 16, 385–398.
- Puyam, A.; Sharma, S.; Kashyap, P.L. RNA interference- a novel approach for plant disease management. J. Appl. Nat. Sci. 2017, 9, 1612–1618.
- Vaucheret, H.; Vazquez, F.; Crété, P.; Bartel, D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004, 18, 1187–1197.
- Limera, C.; Sabbadini, S.; Sweet, J.B.; Mezzetti, B. New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 2017, 8, 1418.
- Gebremichael, D.E.; Haile, Z.M.; Negrini, F.; Sabbadini, S.; Capriotti, L.; Mezzetti, B.; Baraldi, E. RNA interference strategies for future management of plant pathogenic fungi: Prospects and challenges. Plants 2021, 10, 650.
- Kim, D.; Rossi, J. RNAi mechanisms and applications. BioTechniques 2008, 44, 613–616.
- Knip, M.; Constantin, M.E.; Thordal-Christensen, H. Trans-kingdom cross-talk: Small RNAs on the move. PLoS Genet. 2014, 10, e1004602.
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659.
- Mu, J.; Zhuang, X.; Wang, Q.; Jiang, H.; Deng, Z.B.; Wang, B.; Zhang, L.; Kakar, S.; Jun, Y.; Miller, D.; et al. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 2014, 58, 1561–1573.
- Raimondo, S.; Naselli, F.; Fontana, S.; Monteleone, F.; Lo Dico, A.; Saieva, L.; Zito, G.; Flugy, A.; Manno, M.; Di Bella, M.A.; et al. Citrus limon-derived nanovesicles inhibit cancer cell proliferation and suppress CML xenograft growth by inducing TRAIL-mediated cell death. Oncotarget 2015, 6, 19514–19527.
- Wytinck, N.; Manchur, C.L.; Li, V.H.; Whyard, S.; Belmonte, M.F. dsRNA uptake in plant pests and pathogens: Insights into RNAi-based insect and fungal control technology. Plants 2020, 9, 1780.
- Shih, J.D.; Hunter, C.P. SID-1 is a dsRNA-selective dsRNA-gated channel. RNA 2011, 17, 1057–1065.
- Aizawa, S.; Fujiwara, Y.; Contu, V.R.; Hase, K.; Takahashi, M.; Kikuchi, H.; Kabuta, C.; Wada, K.; Kabuta, T. Lysosomal putative RNA transporter SIDT2 mediates direct uptake of RNA by lysosomes. Autophagy 2016, 12, 565–578.
- Wang, M.; Weiberg, A.; Lin, F.-M.; Thomma, B.P.H.J.; Huang, H.-D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 16151.
- McLoughlin, A.G.; Wytinck, N.; Walker, P.L.; Girard, I.J.; Rashid, K.Y.; de Kievit, T.; Fernando, W.G.D.; Whyard, S.; Belmonte, M.F. Identification and application of exogenous dsRNA confers plant protection against Sclerotinia sclerotiorum and Botrytis cinerea. Sci. Rep. 2018, 8, 7320.
- Nerva, L.; Sandrini, M.; Gambino, G.; Chitarra, W. Double-stranded RNAs (dsRNAs) as a sustainable tool against gray mold (Botrytis cinerea) in grapevine: Effectiveness of different application methods in an open-air environment. Biomolecules 2020, 10, 200.
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog 2016, 12, e1005901.
- Werner, B.T.; Gaffar, F.Y.; Schuemann, J.; Biedenkopf, D.; Koch, A.M. RNA-spray-mediated silencing of Fusarium graminearum AGO and DCL genes improve barley disease resistance. Front. Plant Sci. 2020, 11, 476.
- Marcianò, D.; Ricciardi, V.; Marone Fassolo, E.; Passera, A.; Bianco, P.A.; Failla, O.; Casati, P.; Maddalena, G.; De Lorenzis, G.; Toffolatti, S.L. RNAi of a putative grapevine susceptibility gene as a possible downy mildew control strategy. Front. Plant Sci. 2021, 12, 667319.
- Ruiz-Jiménez, L.; Polonio, Á.; Vielba-Fernández, A.; Pérez-García, A.; Fernández-Ortuño, D. Gene mining for conserved, non-annotated proteins of Podosphaera xanthii identifies novel target candidates for controlling powdery mildews by spray-induced gene silencing. J Fungi 2021, 7, 735.
- Fitzgerald, A.; van Kan, J.A.L.; Plummer, K.M. Simultaneous silencing of multiple genes in the apple scab fungus, Venturia inaequalis, by expression of RNA with chimeric inverted repeats. Fungal Genet. Biol. 2004, 41, 963–971.
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, S.D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129.
- Koeck, M.; Hardham, A.R.; Dodds, P.N. The role of effectors of biotrophic and hemibiotrophic fungi in infection: Effectors of biotrophic fungi. Cell. Microbiol. 2011, 13, 1849–1857.
- Tang, C.; Xu, Q.; Zhao, M.; Wang, X.; Kang, Z. Understanding the lifestyles and pathogenicity mechanisms of obligate biotrophic fungi in wheat: The emerging genomics era. Crop. J. 2018, 6, 60–67.
- Braun, U. The current systematics and taxonomy of the powdery mildews (Erysiphales): An overview. Mycoscience 2011, 52, 210–212.
- Vielba-Fernández, A.; Polonio, Á.; Ruiz-Jiménez, L.; de Vicente, A.; Pérez-García, A.; Fernández-Ortuño, D. Fungicide resistance in powdery mildew fungi. Microorganisms 2020, 8, 1431.
- Pérez-García, A.; Romero, D.; Fernández-Ortuño, D.; López-Ruiz, F.; De Vicente, A.; Torés, J.A. The powdery mildew fungus Podosphaera fusca (synonym Podosphaera xanthii), a constant threat to cucurbits. Mol. Plant Pathol. 2009, 10, 153–160.
- Eichmann, R.; Hückelhoven, R. Accommodation of powdery mildew fungi in intact plant cells. J. Plant Physiol. 2008, 165, 5–18.
- Tucker, S.L.; Talbot, N.J. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 2001, 39, 385–417.
- Heffer, V.; Powelson, M.L.; Johnson, K.B.; Shishkoff, N. Identification of powdery mildew fungi anno 2006. Plant Heath Instr. 2006.
- Sidhu, G.S. Genetics of plant pathogenic fungi. In Advances in Plant Pathology; Ingram, D.S., Williams, P.H., Eds.; Academic Press: Cambridge, MA, USA, 1988; Volume 6.
- Gadoury, D.M.; Cadle-Davidson, L.; Wilcox, W.F.; Dry, I.B.; Seem, R.C.; Milgroom, M.G. Grapevine powdery mildew (Erysiphe necator): A fascinating system for the study of the biology, ecology and epidemiology of an obligate biotroph: Grapevine powdery mildew. Mol. Plant Pathol. 2012, 13, 1–16.
- Saharan, G.S.; Mehta, N.K.; Meena, P.D. Infection, pathogenesis and disease cycle. In Powdery Mildew Disease of Crucifers: Biology, Ecology and Disease Management; Springer: Singapore, 2019; pp. 95–130.
- Jarvis, W.R.; Gubler, W.D.; Grove, G.G. Epidemiology of Powdery Mildews in Agricultural Pathosystems; Bélanger, R.R., Bushnell, W.R., Dik, A.J., Carver, T.L.W., Eds.; APS Press: St. Paul, MN, USA, 2002.
- Helfer, S. Rust fungi and global change. New Phytol. 2014, 201, 770–780.
- Aime, M.; Toome, M.; McLaughlin, D. Pucciniomycotina. Systematics and Evolution. In The Mycota; Springer: Berlin/Heidelberg, Germany, 2014; Volume 7A.
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430.
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194.
- Lorrain, C.; Gonçalves dos Santos, K.C.; Germain, H.; Hecker, A.; Duplessis, S. Advances in understanding obligate biotrophy in rust fungi. New Phytol. 2019, 222, 1190–1206.
- Hacquard, S.; Petre, B.; Frey, P.; Hecker, A.; Rouhier, N.; Duplessis, S. The poplar-poplar rust interaction: Insights from genomics and transcriptomics. J. Pathog. 2011, 2011, 716041.
- Sohn, K.H.; Lei, R.; Nemri, A.; Jones, J.D. The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell 2008, 19, 4077–4090.
- Stergiopoulos, I.; de Wit, P.J. Fungal effector proteins. Annu. Rev. Phytopathol. 2009, 47, 233–263.
- Polonio, A.; Pérez-García, A.; Martínez-Cruz, J.; Fernández-Ortuño, D.; de Vicente, A. The haustorium of phytopathogenic fungi: A short overview of a specialized cell of obligate biotrophic plant parasites. In Progress in Botany; Cánovas, F.M., Lüttge, U., Risueño, M.C., Pretzsch, H., Eds.; Springer Nature Switzerland AG: Basel, Switzerland, 2020; Volume 82, ISBN 978-3-030-68619-2.
- Nowara, D.; Gay, A.; Lacomme, C.; Shaw, J.; Ridout, C.; Douchkov, D.; Hensel, G.; Kumlehn, J.; Schweizer, P. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 2010, 22, 3130–3141.
- Pliego, C.; Nowara, D.; Bonciani, G.; Gheorghe, D.M.; Xu, R.; Surana, P.; Whigham, E.; Nettleton, D.; Bogdanove, A.J.; Wise, R.P.; et al. Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. Mol. Plant Microbe Interact. 2013, 26, 633–642.
- Martínez-Cruz, J.; Romero, D.; de la Torre, F.N.; Fernández-Ortuño, D.; Torés, J.A.; de Vicente, A.; Pérez-García, A. The functional characterization of Podosphaera xanthii candidate effector genes reveals novel target functions for fungal pathogenicity. Mol. Plant Microbe Interact. 2018, 31, 914–931.
More
