You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Conner Chen and Version 1 by Massimo Ciccozzi.
Avian influenza virus (AIV) poses a significant challenge to poultry production, with negative repercussions for both the economy and public health worldwide. Since January 2003, a total of 868 human cases of AIV H5N1 have been reported from four countries in the Western Pacific Region, as of 9 March 2023. When AIVs are circulating in poultry, there is a risk of sporadic infections and small clusters of human cases due to exposure to infected poultry or contaminated environments.
- avian influenza virus (AIV)
- H5N1
- Hemoagglutinin (HA)
1. Introduction
Avian influenza virus (AIV), a member of the Orthomyxoviridae family, remains a challenge for poultry production, with negative repercussions for both the micro- and the macro-economy and public health around the world. Globally, from January 2003 to 26 January 2023, there were 868 cases of human infection with avian influenza A(H5N1) virus reported from 21 countries. Of these 868 cases, 457 were fatal [1]. Currently, three types of influenza viruses, based on their antigenic differences, have been described: A, B, and C. Types A and B cause the annual influenza epidemics, which have up to 20% of the population sniffling, aching, coughing, and running high fevers. Type C also causes flu, but its symptoms are much less severe. In addition to these three types, influenza D virus was first identified in pigs and then found to be prevalent in livestock [2,3,4,5][2][3][4][5].
Human pathogens predominantly bind to glycosylated proteins characterized by a terminal sialic acid (SA) α-2,6-galactose (Gal) residue, while avian influenza microorganisms, including H5N1 virus, preferentially bind to SA α-2,3-Gal [6]. AIV primarily infects the respiratory tract, and the main clinical manifestations in humans include nasal discharge, high fever, and weight loss due to dehydration. Severe pathogenesis is represented by lung damage due to extensive infiltration of the lung tissue by inflammatory cells [7].
In response to increasing awareness of circulating AIV strains, the World Health Organization (WHO) closely examines viral antigenicity through sentinel laboratories worldwide to select strains for the development of candidate vaccines for pandemic/epidemic preparedness. To date, vaccines are available for H5, and several drugs, such as Oseltamir, Peramivir, and Zanamivir, have been produced and administered to prevent severe cases [8].
AIV cases may increase globally through migrating birds and poultry trade transmission routes [9]. Poultry vaccination is now considered a relevant and effective control measure. However, the accumulation of point mutations in the viral genome may negatively impact the efficacy of poultry vaccination until a correctly matched vaccine is selected, manufactured, and administered in a timely manner [10]. For this reason, active monitoring of circulating strains is required to constantly prevent the likely emergence of novel viral mutants with epidemic/pandemic potential in both the poultry and human populations [10]. In the last century, the most severe pandemic was the “Spanish Influenza”, caused by the H1N1 virus, as described by Sutton in 2018 [11,12][11][12]. After the 1918 pandemic, H1N1 viruses continued to circulate in humans, and these viruses displayed reduced morbidity and mortality. However, viral reassortment between the 1918 H1N1 strain and AVI viruses resulted in two more pandemics related to the Asian Influenza H2N2 and the Hong Kong Influenza caused by H3N2, which were recorded in 1957 and 1968, respectively [9,11,13][9][11][13]. More recently, in the spring of 2009, a new strain of the influenza A virus (H1N1), initially detected in the United States [14], rapidly spread across the world [15]. This novel strain of H1N1 contained unknown combinations of influenza genes not previously identified in animals or humans and was therefore renamed the influenza A (H1N1) pdm09 virus [16,17][16][17]. The strain was markedly distinct from the H1N1 strains known up to that point, and as a consequence, only a small fraction of young people had any pre-existing immunity [18]. Due to the antigenic differences between the (H1N1) pdm09 virus and the circulating H1N1 viruses, the seasonal flu vaccines provided limited protection against this pandemic virus. From 12 April 2009 to 10 April 2010, the CDC estimated 60.8 million cases of H1N1 influenza in the United States alone [19].
2. Avian Influenza Virus: Genomic Epidemiology
The influenza virus genome is composed of 13 kb and encodes 12 proteins (Figure 1): hemagglutinin (HA), neuraminidase (NA), M1 matrix protein (M1), M2 ion channel protein (M2), nucleocapsid protein (NP), nonstructural protein (NS1, NS2), and RNA polymerase complex (PB1, PB2, PA, PB1-F2 and PA-X) [20]. Influenza A viruses are subtyped based on their combination of HA and NA surface glycoproteins, and up to know a total number of 18 HA and 11 NA subtypes have been already described [21,22,23][21][22][23]. HA is a glycoprotein formed by three district regions. To date, more than 16 different subtypes of HA have been identified and recently classified in two distinct groups and four different clades: (i) group 1, which contains the H1 clade (H1, H2, H5, H6, H11, H13, H16) and the H9 clade (H8, H9, H12); and (ii) group 2, which includes the H3 clade (H3, H4, H14) and the H7 clade (H7, H10, H15) [20,24][20][24].
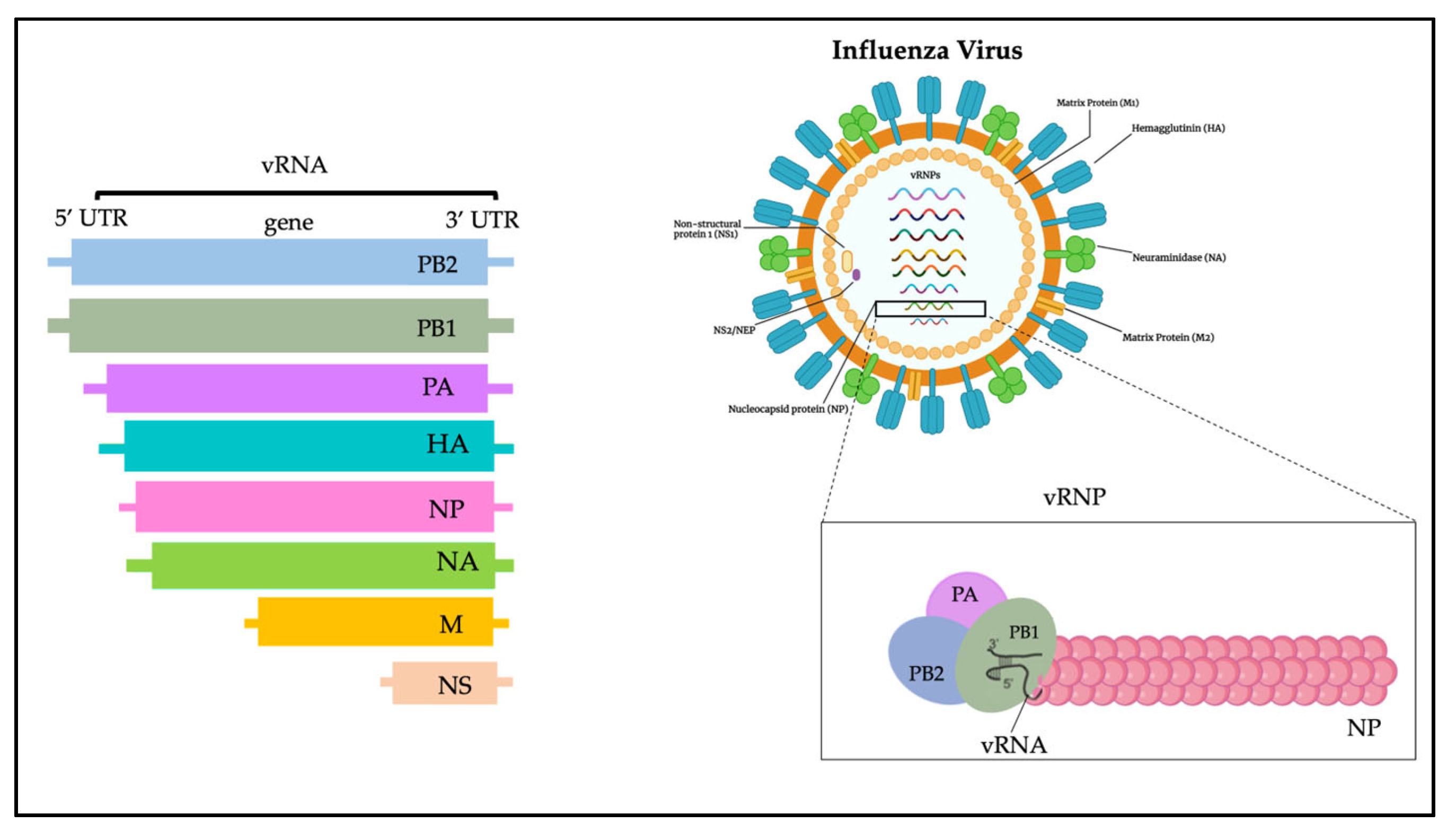
Figure 1.
Influenza virus genome.
NA is instead a tetramer of four identical monomers known as the catalytic head, the stalk, the transmembrane region, and the cytoplasmic tail. NA subtypes are divided into three distinct groups: group 1 (N1, N4, N5, N8); group 2 (N2, N3, N6, N7, N9); and group 3, which includes NA influenza B viruses. N10 and N11 subtypes have been found only in bats [24].
The HA and NA are the prime determinants of the pathogenicity of influenza A viruses. HA attaches virions to cells by binding to terminal sialic acid residues on glycoproteins/glycolipids to initiate the infectious cycle, while NA cleaves terminal sialic acids, releasing virions to complete the infectious cycle [20]. Antibodies specific to HA or NA can protect experimental animals from AIV pathogenesis and drive antigenic variation in their target epitopes that impairs vaccine effectiveness in humans [9,20][9][20].
M1, M2, NS1, and NS2 proteins enhance cytoplasmic trafficking of vesicles, are critical for cytoplasmic trafficking of viral ribonucleoproteins (vRNPs), and aid in the assembly of AIV virions [25]. The M1 protein has a molecular weight of about 26 kDa, and during the last stages of viral replication it translocates into the nucleus and helps to inhibit viral transcription. Subsequently, M1 interacts with the NS2 tail encoded by the viral RNA fragment to expel the vRNPs from the nucleus to the cytoplasm, thus triggering the budding process and the release of virions [26]. The M2 protein is a type III tetrameric integral transmembrane protein and plays an essential role in viral replication by mediating the acidification and shedding of the endosomal trapped virus envelope. This protein is a proton channel activated by low pH; after endocytosis and before HA-mediated fusion between viral and endosomal membrane, M2 channels are activated by low pH of the endosome to conduct protons to acidify the viral interior [27]. Such acidification weakens the electrostatic interaction between ribonucleoprotein (RNP) complexes and matrix proteins. Consequently, membrane fusion can release uncoated RNPs into the cytosol for transport to the nucleus [27].
In the cell replication cycle, NS1 is involved too; it is situated into the nucleus, and its nuclear functions include inhibition of host mRNA processing, blocking of host mRNA nuclear export machinery, and the general inhibition of the host antiviral [28].
NS2 is important in the virus life cycle; it rules the transcription and replication of viral RNA: it is implicated in the regulation of the accumulation of genomic vRNA, antigenomic cRNA and mRNA synthesized by the viral RNA-dependent RNA polymerase. Furthermore, NS2 has been shown to be an important factor in the adaptation of highly pathogenic H5N1 avian influenza viruses to the mammalian host [29].
References
- World Health Organization, WHO. 2023. Available online: https://www.who.int/docs/default-source/wpro---documents/emergency/surveillance/avian-influenza/ai_20230310.pdf?Status=Master&sfvrsn=22ea0816_25 (accessed on 22 March 2023).
- Herfst, S.; Schrauwen, E.J.; Linster, M.; Chutinimitkul, S.; de Wit, E.; Munster, V.J.; Sorrell, E.M.; Bestebroer, T.M.; Burke, D.F.; Smith, D.J.; et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 2012, 336, 1534–1541.
- Ismail, M.M.; Khan, O.A.; Cattoli, G.; Lu, H. Isolation and identification of highly pathogenic avian influenza virus subtype H5N1 in peafowl (Pavo cristatus). Avian Dis. 2010, 54 (Suppl. S1), 357–360.
- Lai, S.; Qin, Y.; Cowling, B.J.; Ren, X.; Wardrop, N.A.; Gilbert, M.; Tsang, T.K.; Wu, P.; Feng, L.; Jiang, H.; et al. Global epidemiology of avian influenza A H5N1 virus infection in humans, 1997–2015: A systematic review of individual case data. Lancet Infect. Dis. 2016, 16, e108–e118.
- Parvin, R.; Begum, J.A.; Nooruzzaman, M.; Chowdhury, E.H.; Islam, M.R.; Vahlenkamp, T.W. Review analysis and impact of co-circulating H5N1 and H9N2 avian influenza viruses in Bangladesh. Epidemiol. Infect. 2018, 146, 1259–1266.
- Sambhara, S.; Poland, G.A. H5N1 Avian influenza: Preventive and therapeutic strategies against a pandemic. Annu. Rev. Med. 2010, 61, 187–198.
- Edenborough, K.M.; Lowther, S.; Laurie, K.; Yamada, M.; Long, F.; Bingham, J.; Payne, J.; Harper, J.; Haining, J.; Arkinstall, R.; et al. Predicting Disease Severity and Viral Spread of H5N1 Influenza Virus in Ferrets in the Context of Natural Exposure Routes. J. Virol. 2015, 90, 1888–1897.
- Li, Y.T.; Linster, M.; Mendenhall, I.H.; Su, Y.C.F.; Smith, G.J.D. Avian influenza viruses in humans: Lessons from past outbreaks. Br. Med. Bull. 2019, 132, 81–95.
- Kanaujia, R.; Bora, I.; Ratho, R.K.; Thakur, V.; Mohi, G.K.; Thakur, P. Avian influenza revisited: Concerns and constraints. Virusdisease 2022, 33, 456–465.
- To, K.K.; Ng, K.H.; Que, T.L.; Chan, J.M.; Tsang, K.Y.; Tsang, A.K.; Chen, H.; Yuen, K.Y. Avian influenza A H5N1 virus: A continuous threat to humans. Emerg. Microbes Infect. 2012, 1, e25.
- Sutton, T.C. The Pandemic Threat of Emerging H5 and H7 Avian Influenza Viruses. Viruses 2018, 10, 461.
- Johnson, N.P.; Mueller, J. Updating the accounts: Global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist. Med. 2002, 76, 105–115.
- Shaw, M.L.; Palese, P. Orthomyxoviridae: The viruses and their replication. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2013; Volume 6, pp. 1691–1740.
- Cohen, J. Swine flu outbreak. Out of Mexico? Scientists ponder swine flu’s origins. Science 2009, 324, 700–702.
- Butler, D. Swine flu goes global. Nature 2009, 458, 1082–1083.
- Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team; Dawood, F.S.; Jain, S.; Finelli, L.; Shaw, M.W.; Lindstrom, S.; Garten, R.J.; Gubareva, L.V.; Xu, X.; Bridges, C.B.; et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 2009, 360, 2605–2615.
- Garten, R.J.; Davis, C.T.; Russell, C.A.; Shu, B.; Lindstrom, S.; Balish, A.; Sessions, W.M.; Xu, X.; Skepner, E.; Deyde, V.; et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009, 325, 197–201.
- Available online: https://www.cdc.gov/flu/pandemic-resources/2009-h1n1-pandemic.html (accessed on 20 March 2023).
- Available online: https://www.who.int/ (accessed on 20 March 2023).
- Shao, W.; Li, X.; Goraya, M.U.; Wang, S.; Chen, J.L. Evolution of Influenza A Virus by Mutation and Re-Assortment. Int. J. Mol. Sci. 2017, 18, 1650.
- Peiris, J.S.; Yu, W.C.; Leung, C.W.; Cheung, C.Y.; Ng, W.F.; Nicholls, J.M.; Ng, T.K.; Chan, K.H.; Lai, S.T.; Lim, W.L.; et al. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 2004, 363, 617–619.
- Fouchier, R.A.; Munster, V.; Wallensten, A.; Bestebroer, T.M.; Herfst, S.; Smith, D.; Rimmelzwaan, G.F.; Olsen, B.; Osterhaus, A.D. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 2005, 79, 2814–2822.
- Webster, R.G.; Govorkova, E.A. Continuing challenges in influenza. Ann. N. Y. Acad. Sci. 2014, 1323, 115–139.
- Tong, S.; Li, Y.; Rivailler, P.; Conrardy, C.; Castillo, D.A.; Chen, L.M.; Recuenco, S.; Ellison, J.A.; Davis, C.T.; York, I.A.; et al. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. USA 2012, 109, 4269–4274.
- Chauhan, R.P.; Gordon, M.L. An overview of influenza A virus genes, protein functions, and replication cycle highlighting important updates. Virus Genes 2022, 58, 255–269.
- Sun, H.; Wu, G.; Zhang, J.; Wang, Y.; Qiu, Y.; Man, H.; Zhang, G.; Li, Z.; Yue, Y.; Tian, Y. Characterization of an intracellular humanized single-chain antibody to matrix protein (M1) of H5N1 virus. PLoS ONE 2022, 17, e0266220.
- Rungrotmongkol, T.; Yotmanee, P.; Nunthaboot, N.; Hannongbua, S. Computational studies of influenza A virus at three important targets: Hemagglutinin, neuraminidase and M2 protein. Curr. Pharm. Des. 2011, 17, 1720–1739.
- Mok, B.W.; Liu, H.; Chen, P.; Liu, S.; Lau, S.Y.; Huang, X.; Liu, Y.C.; Wang, P.; Yuen, K.Y.; Chen, H. The role of nuclear NS1 protein in highly pathogenic H5N1 influenza viruses. Microbes Infect. 2017, 19, 587–596.
- Paterson, D.; Fodor, E. Emerging roles for the influenza A virus nuclear export protein (NEP). PLoS Pathog. 2012, 8, e1003019.
More
