Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 3 by Rita Xu.
The sodium-dependent organic anion transporter (SOAT, gene symbol SLC10A6) specifically transports 3′- and 17′-monosulfated steroid hormones, such as estrone sulfate and dehydroepiandrosterone sulfate, into specific target cells. These biologically inactive sulfo-conjugated steroids occur in high concentrations in the blood circulation and serve as precursors for the intracrine formation of active estrogens and androgens that contribute to the overall regulation of steroids in many peripheral tissues.
- SOAT
- SLC10A6
- sulfated steroids
1. Introduction
Over the last two decades, the sodium-dependent organic anion transporter (SOAT, gene symbol SLC10A6) has been well-established as a specific carrier for sulfated steroid hormones in humans, rats, and mice [1][2][3][4]. The SOAT is a transmembrane protein with nine transmembrane domains (TMDs) that acts as a sodium-dependent uptake carrier for compounds such as dehydroepiandrosterone sulfate (DHEAS), estrone sulfate (E1S), or pregnenolone sulfate (PREGS) [2]. These sulfo-conjugated, biologically inactive steroid metabolites occur in quite high concentrations in the blood circulation and serve as precursors for the intracrine formation of active estrogens and androgens that contribute to the overall regulation of steroids in many peripheral tissues [5][6]. As sulfated steroids are negatively charged at physiological pH, uptake carriers such as SOAT are required to import these molecules for subsequent intracellular cleavage of the sulfate group by the steroid sulfatase STS (so-called sulfatase pathway) [6]. Based on this, only target cells that express uptake carriers such as the SOAT, together with STS, and estrogen/androgen receptors can respond to sulfated steroid hormones [7]. Although SOAT expression has been detected in several peripheral tissues, its quantitative contribution to the steroid sulfate uptake and sulfatase pathway in different organs is still not completely clear. Furthermore, the role of the SOAT in disease progression, particularly in steroid-responsive tumors, and its pharmaceutical potential, e.g., for anti-proliferative therapy, require further investigation.
2. The SLC10 Carrier Family
The SOAT is assigned to the solute carrier family 10 (SLC10) based on its high phylogenetic relationship to the other six members of this carrier family [1][8][9]. The SLC10 family was established in the early 1990s when the first two members, the Na+/taurocholate co-transporting polypeptide (NTCP, gene symbol SLC10A1) and the apical sodium-dependent bile acid transporter (ASBT, gene symbol SLC10A2), were identified by expression cloning [10][11]. The NTCP and ASBT are essentially involved in the maintenance of the enterohepatic circulation of bile acids (Figure 1) by mediating sodium-dependent bile acid uptake in the liver and intestine, respectively [9][12]. Consistent with this function, the SLC10 family was formerly referred to as the “sodium/bile acid co-transporter family” [8]. Later, in the early 2000s, five more genes, referred to as SLC10A3-SLC10A7, were identified that phylogenetically belong to this carrier family [9][12]. Among them, Slc10a6/Soat was first cloned from rat adrenal gland mRNA in 2004 [2]. Just a few years later, the human SLC10A6/SOAT homolog was cloned and functionally characterized [1]. Interestingly, the SOAT showed no transport activity for the most physiologically important bile acids such as taurocholic acid and glycocholic acid, which are the typical substrates of the NTCP and ASBT (Table 1). Instead, the SOAT revealed specific transport activity for all physiologically relevant 3′- and 17′-monosulfated steroid hormones [1][3][13][14]. In addition, only two minor bile acid species, namely taurolithocholic acid-3-sulfate (TLCS) and taurolithocholic acid (TLC), were found to be transported by the SOAT [1][14].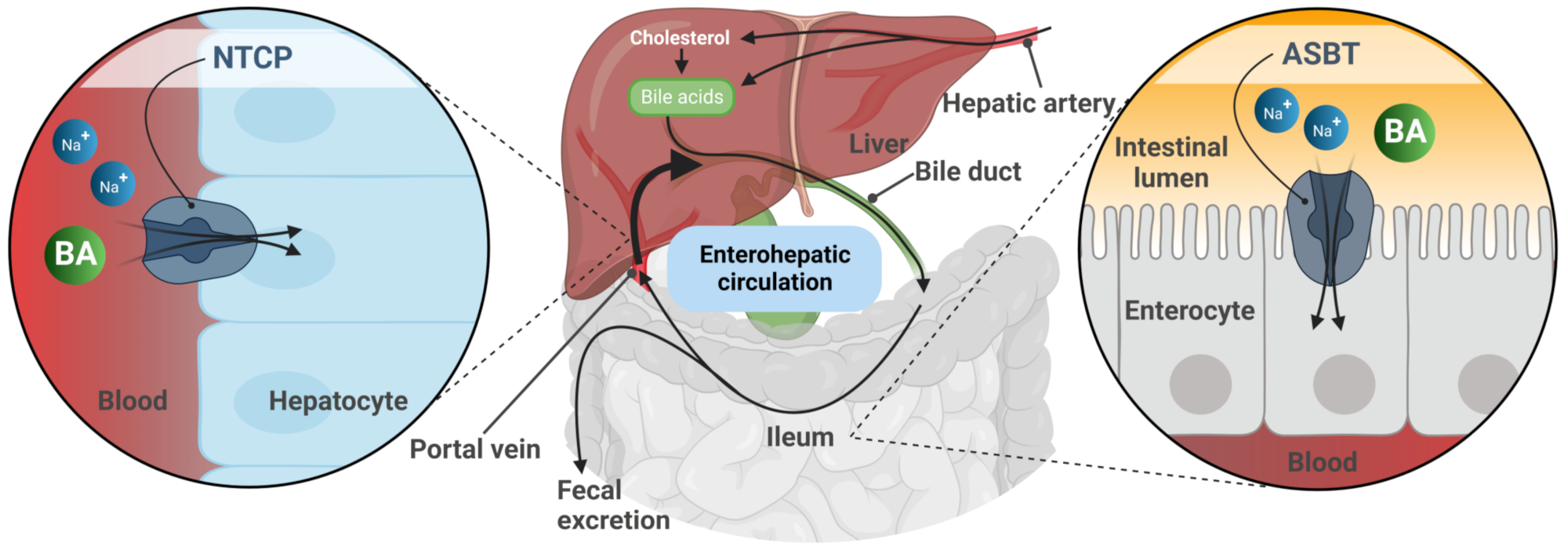
Figure 1. Schematic representation of the enterohepatic circulation of bile acids (BA). After hepatic synthesis and efflux into the bile canaliculi, BA are released into the intestinal lumen and then reabsorbed via the apical sodium-dependent bile acid transporter (ASBT) in the terminal ileum. BA are transported back to the liver with the portal blood flow. Reuptake of BA from portal blood into hepatocytes via the Na+/taurocholate co-transporting polypeptide (NTCP) completes the enterohepatic circulation.
Table 1. Phylogenic relationship of the SLC10 carriers and their substrate patterns.
| Gene | Chromosome | Protein | Protein Length | Transport Substrate | |||
|---|---|---|---|---|---|---|---|
| Steroid Sulfates |
Bile Acids | TLC | |||||
 |
SLC10A1 | 14q24 | NTCP | 349 aa | + | + | + |
| SLC10A4 | 4p11 | P4 | 437 aa | − | − | − | |
| SLC10A2 | 13q33 | ASBT | 348 aa | − | + | + | |
| SLC10A6 | 4q21 | SOAT | 377 aa | + | − | + | |
| SLC10A5 | 8q21 | P5 | 438 aa | − | − | − | |
| SLC10A3 | Xq28 | P3 | 477 aa | − | − | − | |
| SLC10A7 | 4q31 | RCAS | 340 aa | − | − | − | |
3. Genomic Organization of the SLC10A6/Slc10a6 Genes
Phylogenetically, the SOAT and ASBT are most closely related and share identical gene structures. All SLC10A6/Slc10a6 and SLC10A2/Slc10a2 (uppercase for humans, lowercase for animals) genes identified to date have six coding exons with highly conserved exon/intron boundaries, suggesting that both of them share a common ancestor gene [9]. In contrast, the human SLC10A1, SLC10A4, SLC10A5, and SLC10A7 genes exhibit different gene structures with five, three, one, and 11/12 coding exons, respectively. The open reading frame of the human SLC10A6 transcript consists of 1134 base pairs (Figure 2A) and codes for the 377-amino-acid (aa) SOAT protein [1] (Figure 2B). The Soat proteins in rats and mice are slightly shorter, exhibiting 370 and 373 aa, respectively [2][4].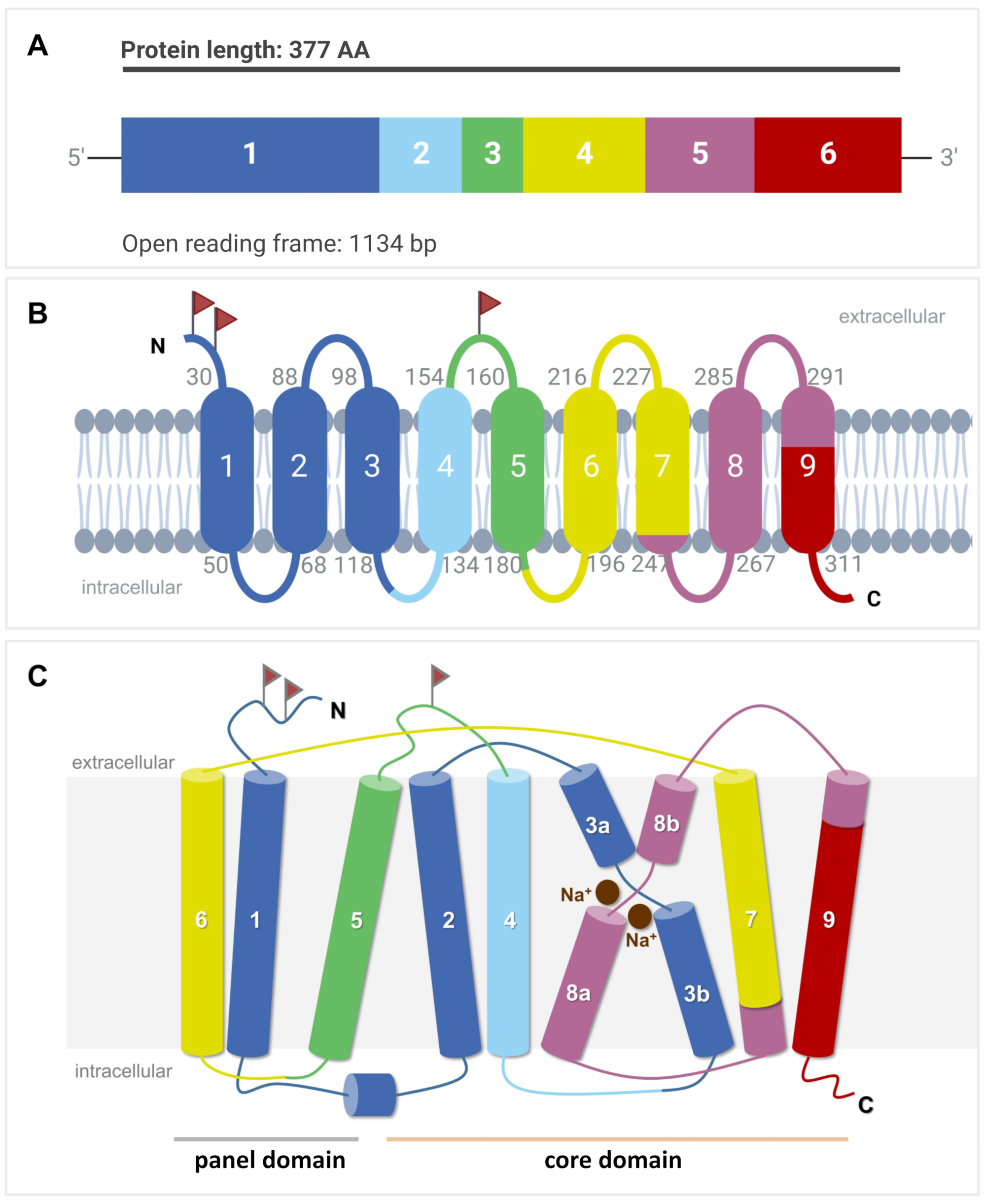
Figure 2. SLC10A6 mRNA transcript and SOAT membrane topology. (A) The open reading frame (ORF) of the SLC10A6 transcript consists of 1134 base pairs (bp), which derive from six coding exons. This ORF is coding for the 377-amino-acid SOAT protein. (B) Schematic transmembrane topology model of the human SOAT protein with nine TMDs and Nexo/Ccyt orientation of the N- and C-terminal ends. The color code indicates which part of the protein is encoded by which corresponding exon. Small gray numbers indicate the amino acid positions of the respective TMD. Red flags indicate possible N-glycosylation sites. (C) Structural arrangement of the TMDs in the core and panel domains. TMDs are numbered from 1 to 9. TMDs 3 and 8 are discontinuous and are named 3a/3b and 8a/8b, respectively. Two Na+ ions are indicated at the proposed sodium-binding sites.
4. SOAT Protein Structure, Sorting, and Dimerization
Based on the crystal structures of two bacterial SLC10-homologous proteins from Neisseria meningitidis and Yersinia frederiksenii named ASBTNm and ASBTYf, respectively [28][29], and the more recent cryogenic electron microscopy (cryo-EM) structures of human NTCP [30][31][32][33], the SOAT is suggested to have a nine-TMD structure with Nexo/Ccyt orientation (Figure 2B). This membrane topology is supported by the de novo structure prediction of the SOAT with AlphaFold [34]. Within this structure, TMDs 1, 5, and 6 form a panel domain, and the other TMDs (2/3/4 and 7/8/9) form a core domain. Within the core domain, TMDs 2/3/4 and 7/8/9 are topologically similar but oppositely orientated within the membrane, revealing an internal twofold pseudosymmetry. TMDs 3 and 8 are discontinuous and cross each other, thereby facilitating the binding of two Na+ ions as the co-substrates (Figure 2C). Substrate binding and translocation occur at the interface between both domains [32]. On the protein level, the SOAT shows the highest sequence homology with the ASBT (42% sequence identity, 70% sequence similarity), followed by the NTCP (33% sequence identity, 63% sequence similarity) [8][9]. The SOAT is a 46 kDa glycoprotein when expressed in HEK293 cells. After PNGase treatment, the apparent molecular weight was found to drop to 42 kDa, most likely representing the non-glycosylated SOAT core protein [1]. There are three potential N-glycosylation sites for the SOAT at amino acid positions Asn4, Asn14, and Asn157, but it is not yet clear where the glycosylation of the SOAT protein exactly occurs (Figure 2B). Sorting studies in transfected HEK293 cells clearly localized the SOAT protein to the plasma membrane [1][13][35][36]. However, immunohistochemistry (IHC) analysis of SOAT expression in different tissues also showed large parts of the protein in the Golgi compartment, at least in primary spermatocytes [13]. Recently, it was shown that homo- and heterodimerization is a common feature of all SLC10 carriers. Different experimental approaches, such as Western blot, co-immunoprecipitation, chemical cross-linking, and membrane-based yeast-two-hybrid screening, have clearly demonstrated the presence of homodimers for the SOAT, NTCP, and ASBT [37][38][39][40][41]. For all three proteins, dimerization seems to be relevant for their regulation and proper expression at the plasma membrane. Interestingly, heterodimerization between different SLC10 carriers has also been demonstrated (e.g., for NTCP/SOAT, NTCP/ASBT, ASBT/SOAT, or SLC10A4/SOAT), suggesting the presence of highly conserved dimerization domains in all SLC10 proteins [41].5. SOAT Substrate Docking and Proposed Transport Mechanism
Very recently, cryo-EM structures of the human NTCP protein were independently solved by four research groups. These studies used detergent-solubilized recombinant NTCP or recombinant NTCP reconstituted in nanodiscs for high-resolution cryo-EM [30][31][32][33]. Essentially, three different conformations of the NTCP protein have been identified: inward-open conformation (PDB 7pqg [31]), outward-open conformation (PDB 7wsi [30], PDB 7fci [33], and PDB 7vad [30]), and open-pore conformation (PDB 7pqq [31] and PDB 7zyi [32]). Due to the high sequence homology between the NTCP and the SOAT, it is assumed that there are also similar protein conformations for the SOAT (Figure 3).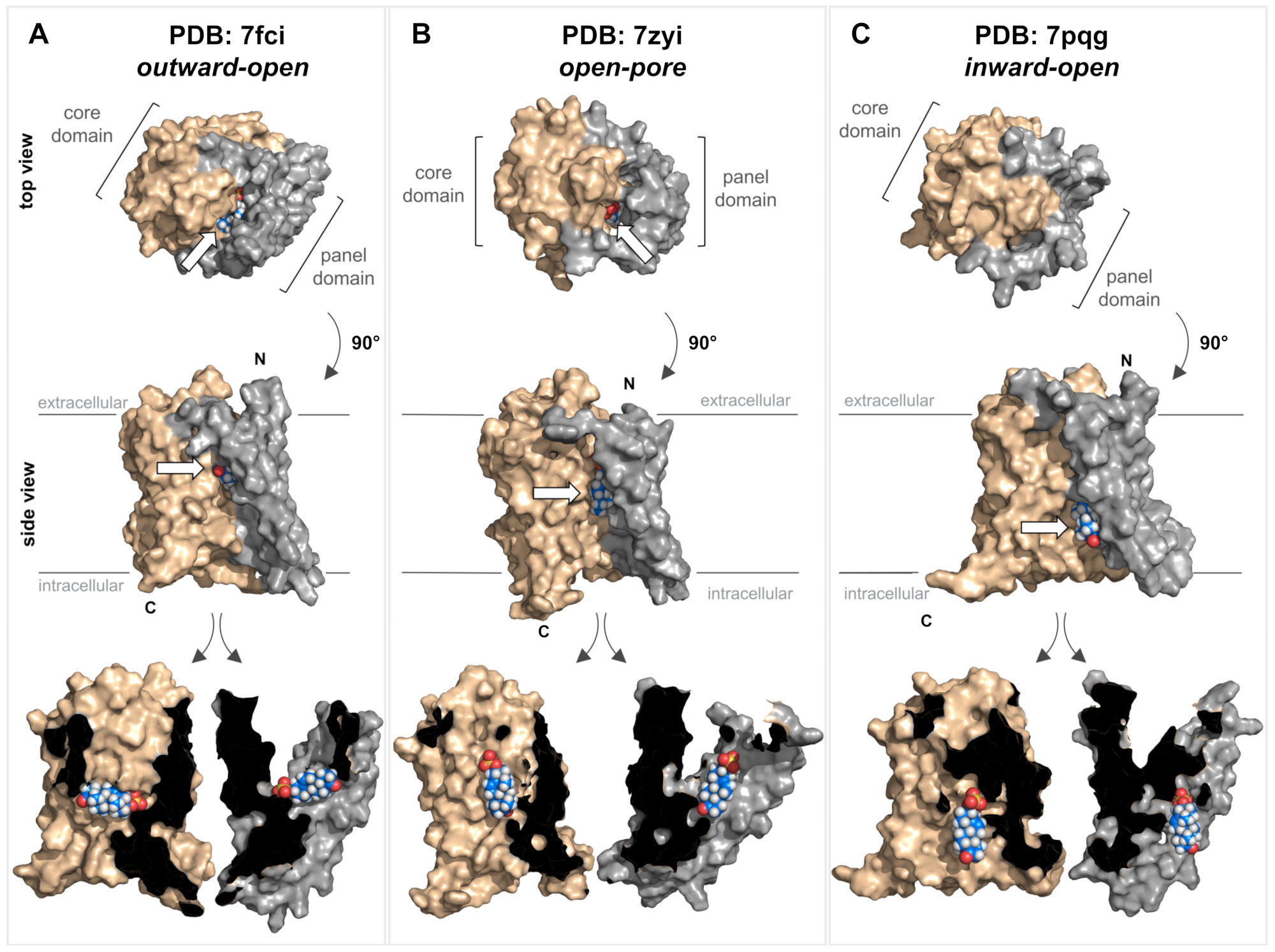
Figure 3. Structure of the SOAT protein and substrate docking. Top and side views of three different homology models of the human SOAT protein (GenBank accession number NP_932069.1) according to SWISS-MODEL predictions. The models were generated based on the recent cryo-EM structures of human NTCP and represent three different conformations: outward-open conformation (A) PDB: 7fci; open-pore conformation (B) PDB: 7zyi; and inward-open conformation (C) PDB: 7pqg. In all the models, the core domain is shown in beige and the panel domain is shown in gray. All structures are shown in both top and side views. In addition, the side-view structures were virtually cut between the core and panel domains, and each was turned outwards to visualize the core/panel interface. All three homology models of human SOAT were used for in silico docking of the SOAT substrate DHEAS (indicated by white arrows). Briefly, the identification of potential binding sites was conducted using the Schrödinger SiteMap program and the receptor grid was set to the interface between the core and the panel domain. The DHEAS structure (downloaded from PubChem at pubchem.ncbi.nlm.nih.gov) was prepared for docking using the Schrödinger LigPrep program. Afterward, the DHEAS molecule was docked to the SOAT protein using the Schrödinger Glide XP program with default settings. All the structures were visualized with PyMOL (version 2.5.4). N, N-terminus; C, C-terminus.
6. SOAT Substrate Recognition
The functional characterization of the SOAT has mostly been carried out in stably transfected Flp-In T-REx HEK293 cells, where the SOAT cDNA was stably integrated at the Flp recognition target (FRT) site, and SOAT expression could be induced by tetracycline treatment [1][3]. Systematic transport studies were performed using different classes of steroids through (I) radiolabeled substrates and liquid scintillation counting (e.g., for [3H]DHEAS, [3H]E1S, [3H]TLC), or (II) liquid chromatography-tandem mass spectrometry (LC-MS/MS) detection of unlabeled compounds (e.g., for 17β-estradiol-3,17-disulfate, 17α-OH-pregnenolone sulfate, testosterone sulfate) [1][3]. In these studies, the SOAT proved to be a very specific carrier for all physiological 3′- and 17′-monosulfated steroid hormones (see Table 2). In contrast, non-sulfated steroids (e.g., estrone, DHEA), glucuronidated steroids (e.g., estradiol-17β-D-glucuronide, estrone-3β-D-glucuronide), and disulfated steroids such as 17β-estradiol-3,17-disulfate were not transported [1][3][14]. This indicates that the presence of one single negatively charged sulfate group at the steroid nucleus is required for the SOAT substrates (Figure 4A). Thereby, α- or β-orientations of the sulfate group were accepted at both the 3′ and 17′ positions, as indicated by the substrate pairs androsterone sulfate (3α) and epiandrosterone sulfate (3β), and epitestosterone sulfate (17α) and testosterone sulfate (17β) (Table 2) [1][3][14].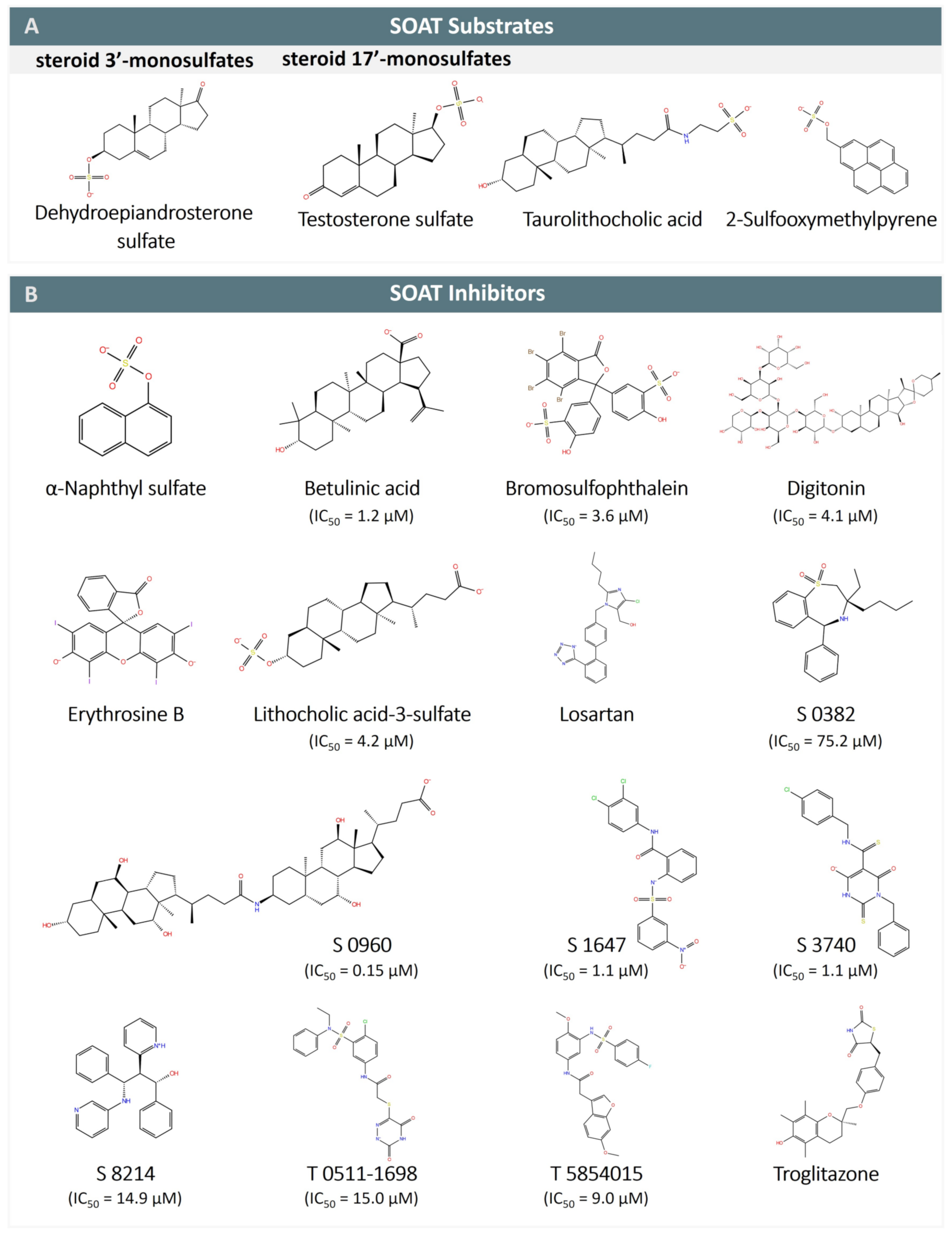
Figure 4. SOAT substrates and inhibitors. (A) SOAT specifically transports steroid 3′- and 17′-monosulfates, represented here by dehydroepiandrosterone sulfate (DHEAS) and testosterone sulfate, respectively. Taurolithocholic acid (TLC) is the only non-sulfated bile acid that is transported by SOAT. 2-sulfooxymethylpyrene (2-SMP) and 4-sulfooxymtehylpyrene (4-SMP) are the only non-steroidal SOAT substrates identified so far. (B) SOAT inhibitors belong to different chemical classes, including steroid-based compounds (bile acids and heart glycosides), organosulfates (α-naphthylsulfate, bromosulfophthalein BSP), phenyl sulfonamides (S 1647, T 0511-1698, and T 5854015), benzothiazepines (S 0382), barbiturates (S 3740), propanolamines (S 8214), thiazolidinediones (troglitazone), and some others. Some of the SOAT inhibitors are also inhibitors of NTCP (e.g., betulinic acid, BSP, erythrosine B, losartan, and troglitazone) and ASBT (e.g., S 0382, S 0960, S 1647, S 3740, S 8214, and troglitazone).
Table 2. SOAT substrates.
| Substrates | Substrate Km |
|---|---|
| Steroid 3’-monosulfates | |
| Pregnenolone sulfate (PREGS) | 11.3 µM [1][3] |
| Estrone sulfate (E1S) | 12.0 µM [1][13] |
| Dehydroepiandrosterone sulfate (DHEAS) | 28.7 µM [1] |
| 16α-OH-DHEAS | 319.0 µM [43] |
| 17α-OH-PREGS | n.d. [3] |
| Androstenediol-3-sulfate | n.d. [13] |
| Androsterone sulfate | n.d. [3] |
| Epiandrosterone sulfate | n.d. [3] |
| 17β-estradiol-3-sulfate | n.d. [13] |
| Steroid 17’-monosulfates | |
| 17β-estradiol-17-sulfate | n.d. [3] |
| 5α-dihydrotestosterone sulfate | n.d. [3] |
| Epitestosterone sulfate | n.d. [3] |
| Testosterone sulfate | n.d. [3] |
| Bile acids | |
| Taurolithocholic acid | 19.3 µM [14] |
| Taurolithocholic acid-3-sulfate | n.d. [1] |
| Non-steroidal organosulfates | |
| 2-Sulfooxymethylpyrene (2-SMP) | n.d. [1] |
| 4-Sulfooxymethylpyrene (4-SMP) | n.d. [1] |
References
- Geyer, J.; Döring, B.; Meerkamp, K.; Ugele, B.; Bakhiya, N.; Fernandes, C.F.; Godoy, J.R.; Glatt, H.; Petzinger, E. Cloning and functional characterization of human sodium-dependent organic anion transporter (SLC10A6). J. Biol. Chem. 2007, 282, 19728–19741.
- Geyer, J.; Godoy, J.R.; Petzinger, E. Identification of a sodium-dependent organic anion transporter from rat adrenal gland. Biochem. Biophys. Res. Commun. 2004, 316, 300–306.
- Grosser, G.; Bennien, J.; Sanchez-Guijo, A.; Bakhaus, K.; Döring, B.; Hartmann, M.; Wudy, S.A.; Geyer, J. Transport of steroid 3-sulfates and steroid 17-sulfates by the sodium-dependent organic anion transporter SOAT (SLC10A6). J. Steroid Biochem. Mol. Biol. 2018, 179, 20–25.
- Grosser, G.; Fietz, D.; Gunther, S.; Bakhaus, K.; Schweigmann, H.; Ugele, B.; Brehm, R.; Petzinger, E.; Bergmann, M.; Geyer, J. Cloning and functional characterization of the mouse sodium-dependent organic anion transporter Soat (Slc10a6). J. Steroid Biochem. Mol. Biol. 2013, 138, 90–99.
- Strott, C.A. Sulfonation and molecular action. Endocr. Rev. 2002, 23, 703–732.
- Mueller, J.W.; Gilligan, L.C.; Idkowiak, J.; Arlt, W.; Foster, P.A. The Regulation of Steroid Action by Sulfation and Desulfation. Endocr. Rev. 2015, 36, 526–563.
- Fietz, D. Transporter for sulfated steroid hormones in the testis—Expression pattern, biological significance and implications for fertility in men and rodents. J. Steroid Biochem. Mol. Biol. 2018, 179, 8–19.
- Döring, B.; Lutteke, T.; Geyer, J.; Petzinger, E. The SLC10 carrier family: Transport functions and molecular structure. Curr. Top. Membr. 2012, 70, 105–168.
- Geyer, J.; Wilke, T.; Petzinger, E. The solute carrier family SLC10: More than a family of bile acid transporters regarding function and phylogenetic relationships. Naunyn Schmiedebergs Arch. Pharmacol. 2006, 372, 413–431.
- Hagenbuch, B.; Meier, P.J. Molecular cloning, chromosomal localization, and functional characterization of a human liver Na+/bile acid cotransporter. J. Clin. Invest. 1994, 93, 1326–1331.
- Wong, M.H.; Oelkers, P.; Dawson, P.A. Identification of a mutation in the ileal sodium-dependent bile acid transporter gene that abolishes transport activity. J. Biol. Chem. 1995, 270, 27228–27234.
- Hagenbuch, B.; Dawson, P. The sodium bile salt cotransport family SLC10. Pflugers Arch. 2004, 447, 566–570.
- Fietz, D.; Bakhaus, K.; Wapelhorst, B.; Grosser, G.; Gunther, S.; Alber, J.; Döring, B.; Kliesch, S.; Weidner, W.; Galuska, C.E.; et al. Membrane transporters for sulfated steroids in the human testis--cellular localization, expression pattern and functional analysis. PLoS ONE 2013, 8, e62638.
- Grosser, G.; Müller, S.F.; Kirstgen, M.; Döring, B.; Geyer, J. Substrate Specificities and Inhibition Pattern of the Solute Carrier Family 10 Members NTCP, ASBT and SOAT. Front. Mol. Biosci. 2021, 8, 689757.
- Claro da Silva, T.; Polli, J.E.; Swaan, P.W. The solute carrier family 10 (SLC10): Beyond bile acid transport. Mol. Aspects Med. 2013, 34, 252–269.
- Burger, S.; Döring, B.; Hardt, M.; Beuerlein, K.; Gerstberger, R.; Geyer, J. Co-expression studies of the orphan carrier protein Slc10a4 and the vesicular carriers VAChT and VMAT2 in the rat central and peripheral nervous system. Neuroscience 2011, 193, 109–121.
- Geyer, J.; Fernandes, C.F.; Döring, B.; Burger, S.; Godoy, J.R.; Rafalzik, S.; Hubschle, T.; Gerstberger, R.; Petzinger, E. Cloning and molecular characterization of the orphan carrier protein Slc10a4: Expression in cholinergic neurons of the rat central nervous system. Neuroscience 2008, 152, 990–1005.
- Schmidt, S.; Moncada, M.; Burger, S.; Geyer, J. Expression, sorting and transport studies for the orphan carrier SLC10A4 in neuronal and non-neuronal cell lines and in Xenopus laevis oocytes. BMC Neurosci. 2015, 16, 35.
- Larhammar, M.; Patra, K.; Blunder, M.; Emilsson, L.; Peuckert, C.; Arvidsson, E.; Ronnlund, D.; Preobraschenski, J.; Birgner, C.; Limbach, C.; et al. SLC10A4 is a vesicular amine-associated transporter modulating dopamine homeostasis. Biol. Psychiatry 2015, 77, 526–536.
- Melief, E.J.; Gibbs, J.T.; Li, X.; Morgan, R.G.; Keene, C.D.; Montine, T.J.; Palmiter, R.D.; Darvas, M. Characterization of cognitive impairments and neurotransmitter changes in a novel transgenic mouse lacking Slc10a4. Neuroscience 2016, 324, 399–406.
- Fernandes, C.F.; Godoy, J.R.; Döring, B.; Cavalcanti, M.C.; Bergmann, M.; Petzinger, E.; Geyer, J. The novel putative bile acid transporter SLC10A5 is highly expressed in liver and kidney. Biochem. Biophys. Res. Commun. 2007, 361, 26–32.
- Godoy, J.R.; Fernandes, C.; Döring, B.; Beuerlein, K.; Petzinger, E.; Geyer, J. Molecular and phylogenetic characterization of a novel putative membrane transporter (SLC10A7), conserved in vertebrates and bacteria. Eur. J. Cell. Biol. 2007, 86, 445–460.
- Ashikov, A.; Abu Bakar, N.; Wen, X.Y.; Niemeijer, M.; Rodrigues Pinto Osorio, G.; Brand-Arzamendi, K.; Hasadsri, L.; Hansikova, H.; Raymond, K.; Vicogne, D.; et al. Integrating glycomics and genomics uncovers SLC10A7 as essential factor for bone mineralization by regulating post-Golgi protein transport and glycosylation. Hum. Mol. Genet. 2018, 27, 3029–3045.
- Dubail, J.; Huber, C.; Chantepie, S.; Sonntag, S.; Tuysuz, B.; Mihci, E.; Gordon, C.T.; Steichen-Gersdorf, E.; Amiel, J.; Nur, B.; et al. SLC10A7 mutations cause a skeletal dysplasia with amelogenesis imperfecta mediated by GAG biosynthesis defects. Nat. Commun. 2018, 9, 3087.
- Laugel-Haushalter, V.; Bar, S.; Schaefer, E.; Stoetzel, C.; Geoffroy, V.; Alembik, Y.; Kharouf, N.; Huckert, M.; Hamm, P.; Hemmerle, J.; et al. A New SLC10A7 Homozygous Missense Mutation Responsible for a Milder Phenotype of Skeletal Dysplasia With Amelogenesis Imperfecta. Front. Genet. 2019, 10, 504.
- Karakus, E.; Wannowius, M.; Müller, S.F.; Leiting, S.; Leidolf, R.; Noppes, S.; Oswald, S.; Diener, M.; Geyer, J. The orphan solute carrier SLC10A7 is a novel negative regulator of intracellular calcium signaling. Sci. Rep. 2020, 10, 7248.
- Wannowius, M.; Karakus, E.; Geyer, J. Functional Analysis of Rare Genetic Variants in the Negative Regulator of Intracellular Calcium Signaling RCAS/SLC10A7. Front. Mol. Biosci. 2021, 8, 741946.
- Hu, N.J.; Iwata, S.; Cameron, A.D.; Drew, D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature 2011, 478, 408–411.
- Zhou, X.; Levin, E.J.; Pan, Y.; McCoy, J.G.; Sharma, R.; Kloss, B.; Bruni, R.; Quick, M.; Zhou, M. Structural basis of the alternating-access mechanism in a bile acid transporter. Nature 2014, 505, 569–573.
- Asami, J.; Kimura, K.T.; Fujita-Fujiharu, Y.; Ishida, H.; Zhang, Z.; Nomura, Y.; Liu, K.; Uemura, T.; Sato, Y.; Ono, M.; et al. Structure of the bile acid transporter and HBV receptor NTCP. Nature 2022, 606, 1021–1026.
- Goutam, K.; Ielasi, F.S.; Pardon, E.; Steyaert, J.; Reyes, N. Structural basis of sodium-dependent bile salt uptake into the liver. Nature 2022, 606, 1015–1020.
- Liu, H.; Irobalieva, R.N.; Bang-Sorensen, R.; Nosol, K.; Mukherjee, S.; Agrawal, P.; Stieger, B.; Kossiakoff, A.A.; Locher, K.P. Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Cell. Res. 2022, 32, 773–776.
- Park, J.H.; Iwamoto, M.; Yun, J.H.; Uchikubo-Kamo, T.; Son, D.; Jin, Z.; Yoshida, H.; Ohki, M.; Ishimoto, N.; Mizutani, K.; et al. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature 2022, 606, 1027–1031.
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444.
- Bakhaus, K.; Fietz, D.; Kliesch, S.; Weidner, W.; Bergmann, M.; Geyer, J. The polymorphism L204F affects transport and membrane expression of the sodium-dependent organic anion transporter SOAT (SLC10A6). J. Steroid Biochem. Mol. Biol. 2018, 179, 36–44.
- Bennien, J.; Fischer, T.; Geyer, J. Rare genetic variants in the sodium-dependent organic anion transporter SOAT (SLC10A6): Effects on transport function and membrane expression. J. Steroid Biochem. Mol. Biol. 2018, 179, 26–35.
- Al-Ansari, N.; Xu, G.; Kollman-Bauerly, K.; Coppola, C.; Shefer, S.; Ujhazy, P.; Ortiz, D.; Ma, L.; Yang, S.; Tsai, R.; et al. Analysis of the effect of intestinal resection on rat ileal bile Acid transporter expression and on bile Acid and cholesterol homeostasis. Pediatr. Res. 2002, 52, 286–291.
- Bijsmans, I.T.; Bouwmeester, R.A.; Geyer, J.; Faber, K.N.; Van de Graaf, S.F. Homo- and hetero-dimeric architecture of the human liver Na(+)-dependent taurocholate co-transporting protein. Biochem. J. 2012, 441, 1007–1015.
- Chothe, P.P.; Czuba, L.C.; Moore, R.H.; Swaan, P.W. Human bile acid transporter ASBT (SLC10A2) forms functional non-covalent homodimers and higher order oligomers. Biochim. Biophys. Acta Biomembr. 2018, 1860, 645–653.
- Kramer, W.; Girbig, F.; Gutjahr, U.; Kowalewski, S.; Jouvenal, K.; Muller, G.; Tripier, D.; Wess, G. Intestinal bile acid absorption. Na(+)-dependent bile acid transport activity in rabbit small intestine correlates with the coexpression of an integral 93-kDa and a peripheral 14-kDa bile acid-binding membrane protein along the duodenum-ileum axis. J. Biol. Chem. 1993, 268, 18035–18046.
- Noppes, S.; Müller, S.F.; Bennien, J.; Holtemeyer, M.; Palatini, M.; Leidolf, R.; Alber, J.; Geyer, J. Homo- and heterodimerization is a common feature of the solute carrier family SLC10 members. Biol. Chem. 2019, 400, 1371–1384.
- Grosser, G.; Baringhaus, K.H.; Döring, B.; Kramer, W.; Petzinger, E.; Geyer, J. Identification of novel inhibitors of the steroid sulfate carrier ‘sodium-dependent organic anion transporter’ SOAT (SLC10A6) by pharmacophore modelling. Mol. Cell. Endocrinol. 2016, 428, 133–141.
- Schweigmann, H.; Sanchez-Guijo, A.; Ugele, B.; Hartmann, K.; Hartmann, M.F.; Bergmann, M.; Pfarrer, C.; Döring, B.; Wudy, S.A.; Petzinger, E.; et al. Transport of the placental estriol precursor 16alpha-hydroxy-dehydroepiandrosterone sulfate (16alpha-OH-DHEAS) by stably transfected OAT4-, SOAT-, and NTCP-HEK293 cells. J. Steroid Biochem. Mol. Biol. 2014, 143, 259–265.
More
