The small intestine has a huge surface area that is further enhanced by villi and microvilli to facilitate the digestion and absorption of nutrients. The expanded surface area of the small intestine increases the likelihood of exposure to pathogens in the lumen. The small intestine must balance the need for nutrient absorption with the ability to ward off pathogens. The majority of the immune cells in the body reside in the mucosa-associated tissues and the mesenchymal tissues of the gastrointestinal tract (GIT). The gut-associated lymphoid tissues (GALT) play a vital role in the development of the immunity of the entire body, as most of the antigens that get into the body are transported to the GIT for processing by its innate immunity before being delivered to the adaptive immunity.
- dysbiosis
- Paneth cells
- immunity
- small intestine
1. Introduction
2. Intestinal Epithelial Cells and Innate Immunity
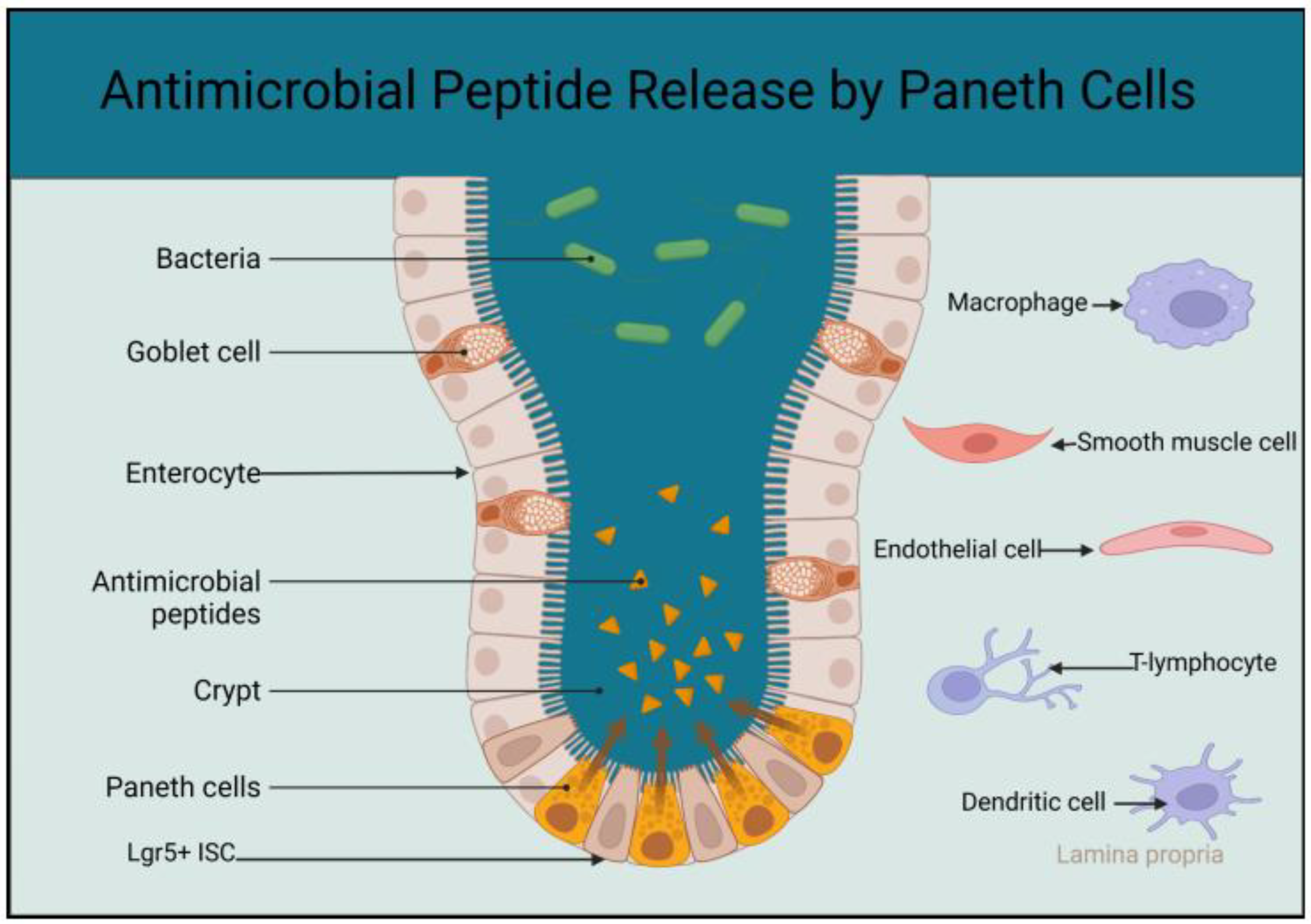
2.1. The Role of Paneth Cells in the Innate Immunity of the Small Intestine
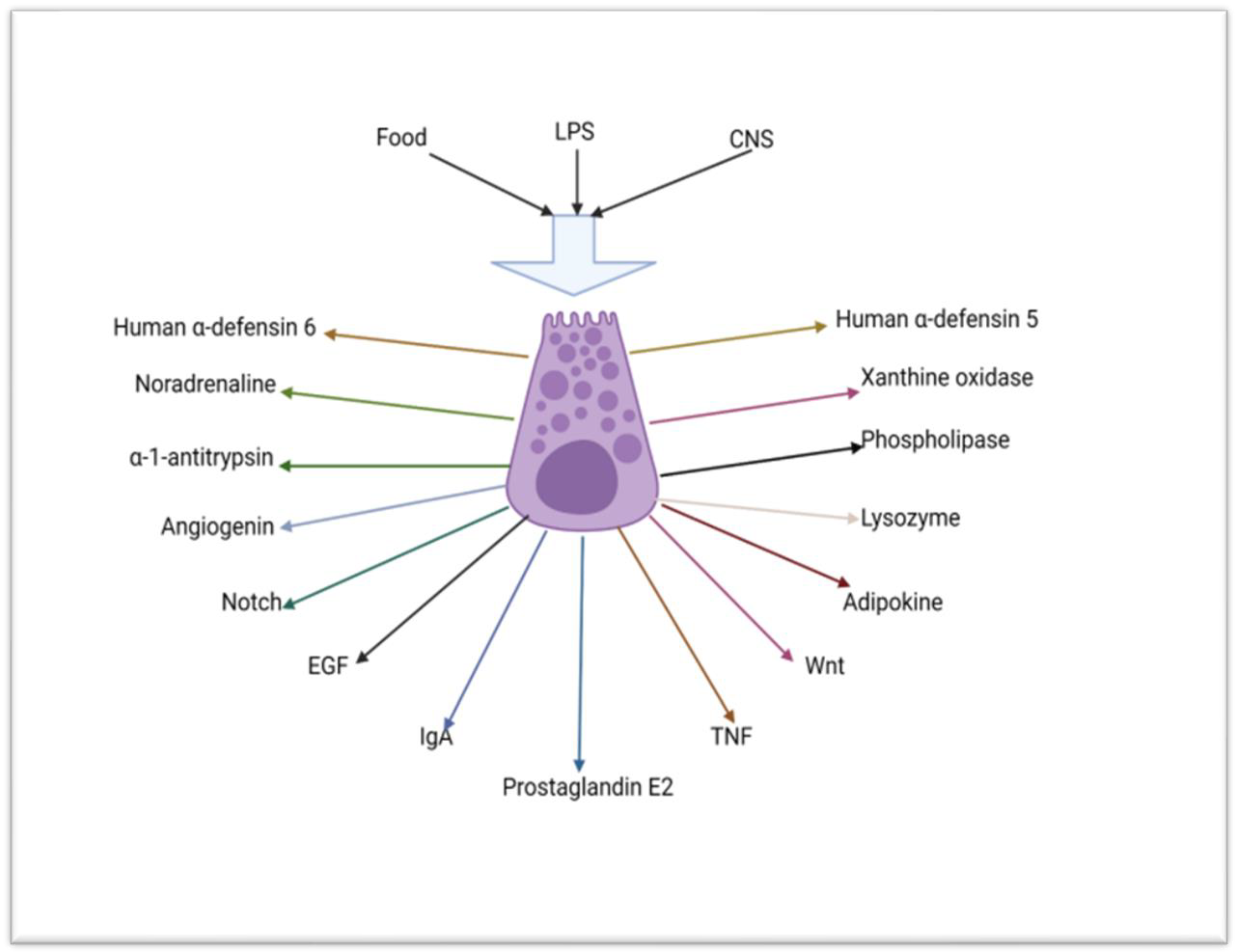

2.2. Absorptive Enterocytes, Goblet Cells, Tuft Cells, M-Cells and Junctional and Innate Immunity of the Small Intestine
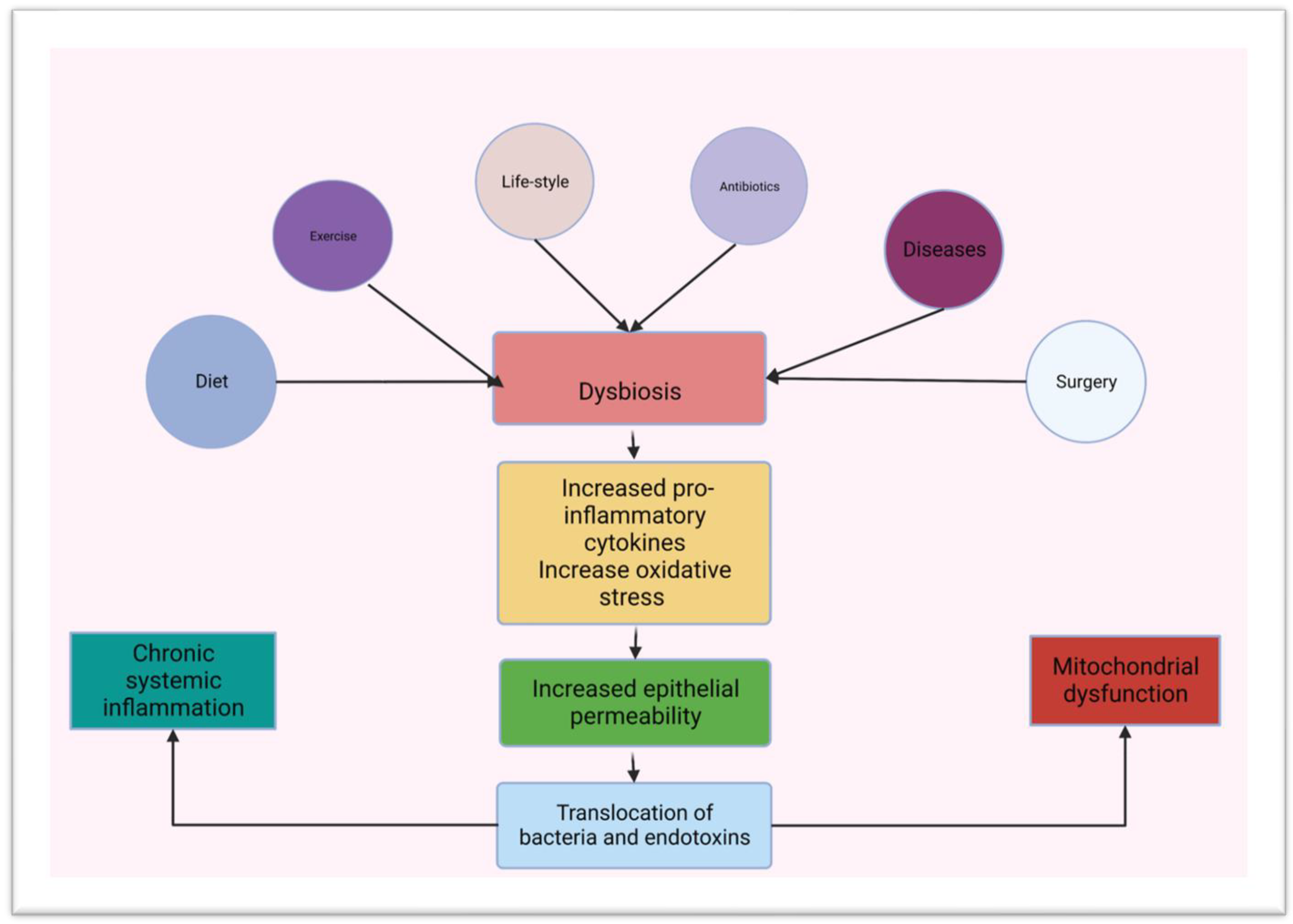
3. Gut Microbiota and Innate Immunity in the Small Intestine
References
- Kobayashi, N.; Takahashi, D.; Takano, S.; Kimura, S.; Hase, K. The Roles of Peyer’s Patches and Microfold Cells in the Gut Immune System: Relevence to Autoimmune Diseases. Front. Immunol. 2019, 10, 2345.
- Basak, O.; van de Born, M.; Korving, J.; Beumer, J.; van der Elst, S.; van Es, J.H.; Clevers, H. Mapping early fate determination in Lgr5+ crypt stem cells using a novel ki67-RFP allele. EMBO J. 2014, 3, 2057–2068.
- Han, H.; Li, Y.; Fang, J.; Liu, G.; Yin, J.; Li, T.; Yin, Y. Gut Microbiota and Type 1 Diabetes. Int. J. Mol. Sci. 2018, 19, 995.
- Clevers, H. The intestinal crypt, a prototype stem cell compartment. Cell 2013, 154, 274–284.
- Beumer, J.; Clevers, H. Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell. Biol. 2020, 22, 39–53.
- Nakamura, K.; Sakuragi, N.; Takakuwa, A.; Ayabe, T. Paneth cell α-defensins and enteric microbiota in health and disease. Biosci. Microbiota Food Health 2016, 35, 57–67.
- Zhu, G.; Hu, J.; Xi, R. The cellular niche for intestinal stem cells: A team effort. Cell Regen. 2021, 10, 1.
- Frost, M.D.; Frank, U.; Weiss, F.U.; Sendler, M.; Kacprowski, T.; Ruhlemann, M.; Bang, C.; Franke, A.; Volker, U.; Volzeke, H.; et al. The Gut Microbiome in Patients with Chronic Pancreatitis Is Characterized by Significant Dysbiosis and Overgrowth by Opportunistic Pathogens. Clin. Transl. Gastroenterol. 2020, 11, e00232.
- Ribeiro, A.B.D.T.M.; Heimesaat, M.M.; Bereswill, S. Changes of the intestinal microbiome-host homeostasis in HIV-infected individuals-a focus on bacterial gut microbiome. Eur. J. Microbiol. Immunol. 2017, 7, 158–167.
- Cari, L.; Rosati, L.; Leoncini, G.; Lusenti, E.; Gentili, M.; Nocentini, C.; Riccardi, C.; Migliorati, G.; Ronchetti, S. Association of GILZ with MUC2, TLR2 and TLR4 in Inflammatory Bowel Disease. Int. J. Mol. Sci. 2023, 24, 2235.
- Salazar, J.; Angarita, L.; Morillo, V.; Navarro, C.; Martinez, M.S.; Chacin, M.; Torres, W.; Rajotia, A.; Rojas, M.; Cano, C.; et al. Microbiota and Diabetes Mellitus: Role of Lipid Mediators. Nutrients 2020, 12, 3039.
- Sharma, S.; Tripathi, P. Gut microbiome and type 2 diabetes: Where we are and where to go? J. Nutr. Biochem. 2018, 63, 101–108.
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503.
- Al-Rashidi, H.E. Gut microbiota and immunity relevance in eubiosis and dysbiosis. Saudi J. Biol. Sci. 2022, 29, 1628–1643.
- Lazar, V.; Ditu, L.M.; Pircalabioru, G.G.; Curutiu, C.; Holban, A.M.; Picu, A.; Petcu, L.; Chifiriuc, M.C. Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Front. Immunol. 2018, 9, 1830.
- Hackam, D.J.; Sodhi, C.P. Toll-Like Receptor-Mediated Intestinal Inflammatory Imbalances in the Pathogenesis of Necrotizing Enterocolitis. Cell. Mol. Gastroenterol. Hepatol. 2018, 6, 229–238.e1.
- Lievin-Le Moal, V.; Servin, A.L. The frontline of enteric host defense against unwelcome intrusion of harmful microorganisms: Mucins, antimicrobial peptides, and microbiota. Clin. Microbiol. Rev. 2006, 19, 315–337.
- Allam-Ndoul, B.; Castonguay-Paradis, S.; Veilleux, A. Gut Microbiota and Intestinal Trans-Epithelial Permeability. Int. J. Mol. Sci. 2020, 21, 6402.
- Mukerjee, S.; Hooper, L.V. Antimicrobial Defense of the Intestine. Immunity 2015, 42, 28.
- Sankaran-Walters, S.; Hart, R.; Dills, C. Guardians of the Gut Enteric Defensins. Front. Microbiol. 2017, 8, 647.
- Bevins, C.L. Events at the host-microbial interface of the gastrointestinal tract. V. Paneth cell alpha-defensins in intestinal host defense. Am. J. Physiol. Liver Physiol. 2005, 289, G173–G176.
- Cray, P.; Sheahan, B.J.; Dekaney, C.M. Secretory Sorcery: Paneth Cell Control of Intestinal Repair and Homeostasis. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 1239–1250.
- Dinsdale, D.; Biles, B. Postnatal changes in the distribution and elemental composition of Paneth cells in normal and corticosteroid-treated rats. Cell Tissue Res. 1986, 246, 183–187.
- Zhang, M.; Wu, C. The relationship between intestinal goblet cells and the immune response. Biosci. Rep. 2020, 40, BSR20201471.
- Singh, R.; Balasubramanian, I.; Zhang, L.; Gao, N. Metaplastic Paneth Cells in Extra-Intestinal Mucosal Niche Indicate a Link to Microbiome and Inflammation. Front. Physiol. 2020, 11, 280.
- Lueschow, S.R.; McElroy, S.J. The Paneth Cell: The Curator and Defender of the Immature Small Intestine. Front. Physiol. 2020, 11, 587.
- Lee, V.H.; Gulati, A.S. Implications of Paneth cell dysfunction on gastrointestinal health and disease. Curr. Opin. 2022, 38, 535–540.
- Ma, Q.; Li, Y.; Li, P.; Wang, M.; Wang, J.; Tang, Z.; Wang, T.; Luo, L.; Wang, C.; Wang, T.; et al. Research progress in the relationship between type 2 diabetes mellitus and intestinal flora. Biomed. Pharmacother. 2019, 117, 109138.
- Grinat, J.; Kosel, F.; Goveas, N.; Kranz, A.; Alexopoulou, D.; Rajewsky, K.; Sigal, M.; Stewart, A.F.; Heuberger, J. Epigenetic modifier balances Mapk and Wnt signalling in differentiation of goblet and Paneth cells. Life Sci. Alliance 2022, 5, e202101187.
- Holly, M.K.; Smith, J.G. Paneth Cells during Viral Infection and Pathogenesis. Viruses 2018, 10, 225.
- Salzman, N.H.; Underwood, M.A.; Bevins, C.L. Paneth cells, defensins, and the commensal microbiota: A hypothesis on intimate interplay at the intestinal mucosa. Semin. Immunol. 2007, 19, 70–83.
- Farin, H.F.; Karthaus, W.R.; Kujala, P.; Rakhshandehroo, M.; Schwank, G.; Vries, R.G.J.; Kalkhoven, E.; Nieuwenhuis, E.E.S.; Clevers, H. Paneth cell extrusion and release of antimicrobial products is directly controlled by immune cell-derived IFN-γ. J. Exp. Med. 2014, 211, 1393–1405.
- Ouellette, A.J. Paneth cell α-defensins in enteric immunity. Cell. Mol. Life Sci. 2011, 68, 2215–2229.
- Gassler, N. Paneth cells in intestinal physiology and pathophysiology. World J. Gastrointest. Pathophysiol. 2017, 8, 150–160.
- Chairatana, P.; Nolan, E.M. Human α-Defensin 6: A Small Peptide that Self-Assembles and Protects the Host by Entangling Microbes. Acc. Chem. Res. 2017, 50, 960–967.
- Dayton, T.L.; Clevers, H. Beyond growth signaling: Paneth cells metabolically support ISCs. Cell Res. 2017, 27, 851–852.
- Fruitwala, S.; El-Naccache, D.W.; Chang, T.L. Multifaceted immune functions of human defensins and underlying mechanisms. Semin. Cell Dev. Biol. 2019, 88, 163–172.
- Hein, M.J.A.; Kvansakul, M.; Lay, F.T.; Phan, T.K.; Hulett, M.D. Defensin-lipid interactions in membrane targeting: Mechanisms of action and opportunities for the development of antimicrobial and anticancer therapeutics. Biochem. Soc. Trans. 2022, 50, 423–437.
- Gao, X.; Ding, J.; Liao, C.; Xu, J.; Liu, X.; Lu, W. Defensins: The natural peptide antibiotic. Adv. Drug Deliv. Rev. 2021, 179, 114008.
- Jackson, D.; Theiss, A.L. Intestinal Stem Cell Regulation via Glycolytic Activity of Neighboring Paneth Cells. J. Gastroenterol. Hepatol. Endosc. 2017, 2, 1019.
- Heneghan, A.F.; Pierre, J.F.; Tandee, K.; Shanmuganayagam, D.; Wang, X.; Reed, J.D.; Steele, J.L.; Kudsk, K.A. Parenteral Nutrition Decreases Paneth cell function and Intestinal Bactericidal Activity while Increasing Susceptibility to Bacterial Enteroinvasion. J. Parenter. Enter. Nutr. 2014, 38, 817–824.
- Hodin, C.M.; Lenaerts, K.; Grootjans, J.; de Haan, J.J.; Hadfoune, M.; Verheyen, F.K.; Kiyama, H.; Heineman, E.; Buurman, W.A. Starvation Compromises Paneth Cells. Am. J. Pathol. 2011, 179, 2885.
- Yang, E.; Shen, J. The Roles and functions of Paneth cells in Crohn’s disease: A critical review. Cell Prolif. 2020, 54, e12958.
- Liu, T.C.; Gurram, B.; Baldridge, M.T.; Head, R.; Lam, V.; Luo, C.; Cao, Y.; Simpson, P.; Hayward, M.; Holtz, M.L.; et al. Paneth cell defects in Crohn’s disease patients promotes dysbiosis. J. Clin. Investig. 2016, 1, e86907.
- Liu, T.C.; Kern, J.T.; VanDussen, K.L.; Xiong, S.; Kaiko, G.E.; Wilen, C.B.; Rajala, M.W.; Caruso, R.; Holtzman, M.J.; Gao, F.; et al. Interaction between smoking and ATG16L1T300A triggers Paneth cell defects in Crohn’s disease. J. Clin. Investig. 2018, 128, 5110–5122.
- Stappenbeck, T.S.; McGovern, D.P.B. Paneth Cell Alterations in the Development and Phenotype of Crohn’s Disease. Gastroenterology 2017, 152, 322–326.
- Donaldson, D.S.; Shih, B.B.; Mabbott, N.A. Aging-Related Impairments to M Cells in Peyers’s Patches Coincide with Disturbances to Paneth Cells. Front. Immunol. 2021, 12, 761949.
- Kelly, P.; Feakins, R.; Domizio, P.; Murphy, J.; Bevins, C.; Wilson, J.; McPhail, G.; Poulsom, R.; Dhaliwal, W. Paneth cell granule depletion in the human intestine under infective and nutrional stress. Clin. Exp. Immunol. 2004, 135, 303–309.
- Latorre, E.; Layunta, E.; Grasa, L.; Pardo, J.; Garcia, S.; Alcalde, A.I.; Mesonero, J.E. Toll-like receptots 2 and 4 modulate intestinal IL-10 differently in ileum and colon. United Eur. Gastroenterol. J. 2018, 6, 446–453.
- Kwon, M.S.; Chung, H.K.; Xiao, L.; Yu, T.X.; Wang, S.R.; Piao, J.J.; Rao, J.N.; Gorospe, M.; Wang, J.Y. MicroRNA-195 regulates Tuft cell function in the intestinal epithelium by altering translation of DCLK1. Am. J. Physiol. Cell Physiol. 2021, 320, C1042–C1054.
- Schneider, C.; O’Leary, C.E.; Locksley, R.M. Regulation of immune responses by tuft cells. Nat. Rev. Immunol. 2019, 19, 584–593.
- Billipp, T.E.; Nadjsombati, M.S.; von Moltke, J. Tuning tuft cells: New ligands and effector functions reveal tissue-specific function. Curr. Opin. Immunol. 2021, 68, 98–106.
- O’Leary, C.E.; Schneider, C.; Locksley, R.M. Tuft cells-systemically dispersed sensory epithelia integrating immune and neural circuitry. Annu. Rev. Immuno.l 2019, 37, 47–72.
- Han, S.J.; Kim, M.; D’Agati, V.D.; Lee, H.T. Norepinephrine released by intestinal Paneth cells exacerbates ischemic AKI. Am. J. Physiol. Ren. Physiol. 2020, 318, F260–F272.
- Hills, R.D.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut Microbiome: Profound Implications for Diet and Disease. Nutrients 2019, 11, 1613.
- Chen, Y.; Zhou, J.; Wang, L. Role and Mechanism of Gut in Human Disease. Front. Cell. Infect. Microbiol. 2021, 11, 625913.
- Siljander, H.; Honkanen, J.; Knip, M. Micriobiome and Type 1 diabetes. EBioMedicine 2019, 46, 512–521.
- Ortega, M.A.; Fraile-Martinez, O.; Naya, I.; Garcia-Honduvilla, N.; Alvarez-Mon, M.; Bujan, J.; Asunsolo, A.; de la Torre, B. Type 2 Diabetes Mellitus Associated with Obesity (Diabesity). The Central Role of Gut Microbiota and Its Translational Applications. Nutrients 2020, 12, 2749.
- Stecher, B.; Hardt, W.D. Mechanisms controlling pathogen colonization of the gut. Curr. Opin. Microbiol. 2011, 14, 82–91.
- Khanna, S.; Tosh, P.K. A clinician’s primer of the role of the microbiome in human health and disease. Mayo Clin. Proc. 2014, 89, 107–114.
- Hasain, Z.; Mokhtar, N.M.; Kamaruddin, N.A.; Ismail, N.A.M.; Razalli, N.H.; Gnanou, J.V.; Ali, R.A.R. Gut Microbia and Gestational Daibetes Mellitus: A Review of Host-Gut Microbiota Interactions and Their Therapeutic Potential. Front. Cell. Infect. Microbiol. 2020, 10, 188.
- Cani, P.D. Human gut microbiome: Hopes, threats and promises. Gut 2018, 67, 1716–1725.
- Tsai, Y.L.; Lin, T.L.; Chang, C.J.; Wu, T.R.; Lai, W.F.; Lu, C.C.; Lai, H.C. Probiotics, prebiotics and amelioration of diseases. J. Biomed. Sci. 2019, 26, 3.
- Stern, J.; Miller, G.; Li, X.; Saxena, D. Virome and bacteriome: Two sides of the same coin. Curr. Opin. Virol. 2019, 37, 37–43.
- Yang, K.; Niu, J.; Zuo, T.; Sun, Y.; Xu, Z.; Tang, W.; Liu, Q.; Zhang, J.; Ng, E.K.W.; Wong, S.K.H.; et al. Alterations in the Gut Virome in Obesity and Type 2 Diabetes Mellitus. Gastroenterology 2021, 161, 1257–1269.
- Xu, J.; Xu, X.; Chen, X.; Cai, X.; Yang, S.; Sheng, Y.; Wang, T. Regulation of an antioxidant blend on intestinal redox status and major microbiota in early weaned piglets. Nutrition 2014, 30, 584–589.
- Yokoi, Y.; Nakamura, K.; Yoneda, T.; Kikuchi, M.; Sugimoto, R.; Shimizu, Y.; Ayabe, T. Paneth cell granule dynamics on secretory responses to bacterial stimuli in enteroids. Sci. Rep. 2019, 9, 2710.
- Singh, V.; Ahlawat, S.; Mohan, H.; Gill, S.S.; Sharma, K.K. Balancing reactive oxygen species generation by rebooting gut microbiota. J. Appl. Microbiol. 2022, 132, 4112–4129.
- Jimenez-Uribe, A.P.; Hernandez-Cruz, E.Y.; Ramirez-Magana, K.J.; Pedraza-Chaverri, J.P. Involvement of Tricarboxylic Acid Cycle Metabolites in Kidney Diseases. Biomolecules 2021, 11, 1259.
- Nagpal, R.; Newman, T.M.; Wang, S.; Jain, S.; Lovato, J.F.; Yadav, H. Obesity-Linked Gut Microbiome Dysbiosis Associated with Derangements in Gut Permeability and Intestinal Cellular Homeostasis Independent of Diet. J. Diabetes Res. 2018, 2018, 3462092.
- Zhou, Z.; Sun, B.; Yu, D.; Zhu, C. Gut Microbiota: An Important Player in Type 2 Diabetes Mellitus. Front. Cell. Infect. Microbiol. 2022, 12, 834485.
- Singh, R.B.; Fedacko, J.; Fatima, G.; Magomedova, A.; Watanabe, S.; Elkilany, G. Why and How the Indo-Mediterranean Diet May Be Superior to Other Diets: The Role of Antioxidants in the Diet. Nutrients 2022, 14, 898.
- Darenskaya, M.A.; Kolesnikova, L.I.; Kolesnikov, S.I. Oxidative Stress: Pathogenetic Role in Diabetes in Diabetes Mellitus and Its Complications and Therapeutic Approaches to Correction. Bull. Exp. Biol. Med. 2021, 171, 179–189.
- Karam, B.S.; Chavez-Moreno, A.; Koh, W.; Akar, J.G.; Akar, F.G. Oxidative stress and inflammation as central mediators of atrial fibrillation in obesity and diabetes. Cardiovasc. Diabetol. 2017, 16, 120.
- Shan, Z.; Fa, W.H.; Tian, C.R.; Yuan, C.S.; Jie, N. Mitophagy and mitochondrial dynamics in type 2 diabetes mellitus treatment. Aging 2022, 14, 2902–2919.
- Berger, N.A. Young Adult Cancer: Influence of the Obesity Pandemic. Obesity 2018, 26, 641–650.
- Capuco, A.; Urits, I.; Hasoon, J.; Chun, R.; Gerald, B.; Wang, J.K.; Kassem, H.; Ngo, A.L.; Abd-Elsayed, A.; Simopoulos, T.; et al. Current Perspectives on Gut Microbiome Dysbiosis and Depression. Adv. Ther. 2020, 37, 1328–1346.
- Khandekar, M.J.; Cohen, P.; Spiegelman, B.M. Molecular mechanisms of cancer development in obesity. Nat. Rev. Cancer 2011, 11, 886–895.
- Kawai, T.; Auteieri, M.V.; Scalia, R. Adipose tissue inflammation and metabolic dysfunction in obesity. Am. J. Physiol. 2021, 320, C375–C391.
- Tao, Z.; Shi, A.; Zhao, J. Epidemiological Perspectives of Diabetes. Cell Biochem. Biophys. 2015, 73, 181–185.
- Pircalabioru, G.G.; Corcionivoschi, N.; Gundogdu, O.; Chifiriuc, M.C.; Marutescu, L.G.; Ispas, B.; Savu, O. Dysbiosis in the Development of Type! Diabetes and Associated Complications: From Mechanisms to Targeted Gut Microbes Manipulation Therapies. Int. J. Mol. Sci. 2021, 22, 2763.
- Iyengar, N.M.; Gucalp, A.; Dannenberg, A.J.; Hudis, C.A. Obesity and Cancer Mechanisms: Tumor Microenvironment and Inflammation. J. Clin. Oncol. 2016, 34, 4270–4276.
