Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Alessandro Fantoni and Version 2 by Jessie Wu.
The progresses in the research of plasmonic phenomena and materials paved the route toward the development of optical sensing platforms based on metal nanostructures with a great potential to be integrated into point-of-care (POC) devices for the next generation of sensing platforms, thus enabling real-time, highly sensitive and accurate diagnostics.
- POC devices
- plasmonic biosensors
- readout systems
- multiplexed detection
1. Smartphone-Assisted Devices
In the work of Choi [1][60], a smartphone-assisted point-of-care (POC) bioassay to detect urea for the daily monitoring of renal and hepatic dysfunction diseases was developed. At the basis of the sensing system, there is the colorimetric detection of urea relying on the production of silver nanoparticles (AgNPs) integrated with an image-analysis system by means of a smartphone. Through urease, urea is hydrolyzed in NH3 and CO2, thus activating tannic acid, which is sensitive to pH, and reducing Ag+ ions into AgNPs. Due to their LSPR, AgNPs display a yellow colour: the quality of the colour, associated with the LSPR position and intensity, depends on the number of particles generated. To facilitate the readout of the signal, a paper sheet was exploited as the substrate: using a laser printer, circular hydrophobic black zones were printed around hydrophilic sampling areas. The hydrophilic area inside the black hydrophobic circles was then loaded with urease; in a successive step, solutions at different concentrations of urea were added to the areas of the paper strips functionalized with urease. After a few minutes, the sensing solution composed of tannic acid and AgNO3 was added and, as a result, the sensing area displayed several colours depending on the urea concentration. In this direction, a rough analysis of the urea concentration is possible with the naked eye, but to obtain more precise information, an analysis assisted by a smartphone was performed. The corrected RGB ratio showed a linear correlation with urea concentration, so that the urea concentration was determined using smartphone software based on that relation, obtaining an LOD of 2168 mM for urea in human urine samples. Stimulated by the need for a fast and reliable test, the ironPhone system was developed in the work of Srinivasan [2][61] as a POC device to quantify human serum-ferritin concentrations, a biomarker for the status of iron. The ironPhone system is composed of a lateral flow immunoassay (LFIA) test strip developed specifically for serum ferritin, a test-strip reader and the ironPhone app to display the test results after analysis realization. The first part of the sensing device is the LFIA test strip contained in a cartridge and consisting of a polyester backing layer on which a sample, conjugation and absorption pad are deposited along with a nitrocellulose membrane containing the test and the control line. The LFIA test strip was designed to perform a sandwich immunoassay analysis: firstly, a drop of blood from a finger is introduced onto the inlet of the test strip, in the sample pad, followed by the chase buffer, saturating the sample pad and thus initiating the capillary flow needed to transport the elements of the sandwich immunoassay. In the sample pad, the whole blood sample is filtered, so that the serum is separated and it alone is allowed to flow along the conjugation pad. After the filtration, in the conjugation pad, the serum interacts with the AuNP–anti-ferritin conjugates, which are able to bind the ferritin molecules contained in the initial sample. Successively, the AuNP–anti-ferritin conjugates transporting the ferritin molecules flow to the nitrocellulose membrane, which binds to the polyclonal anti-ferritin antibody forming the test line, thus creating a strong colorimetric signal. The AuNP–anti-ferritin conjugates passing through the test line without binding are then captured by the anti-mouse antibody composing the control line, thus displaying a weak colorimetric signal: as the ferritin concentration increases, the colour intensity of the control line decreases; whereas, for lower ferritin levels, the test line shows a weak colour leading to a strong signal in the control line, indicating that the most of the unbound AuNP–anti-ferritin is bound to the secondary antibody of the control line. At this point, the sample flows to the absorbent pad. Once that the test and control line are ready and the sample has run until the absorption pad, the test strip is inserted within the LFIA strip reader, where the colorimetric signals are amplified and captured from a camera and analysed using the ironPhone app to obtain information about the ferritin concentration. In 2016, the same group developed the NutriPhone system, a system comprising of a test strip, a test-strip reader and a smartphone app, to quantify the levels of Vitamin B12 in the human blood [3][62]. NutriPhone works in the same way of the ironPhone system, but, in this case, the AuNP–anti-ferritin conjugates were replaced with AuNP–anti-B12 conjugates and the test and control lines were composed by Vitamin B12-BSA (bovine serum albumin) conjugate and anti-mouse IgG produced in goat (Figure 1).
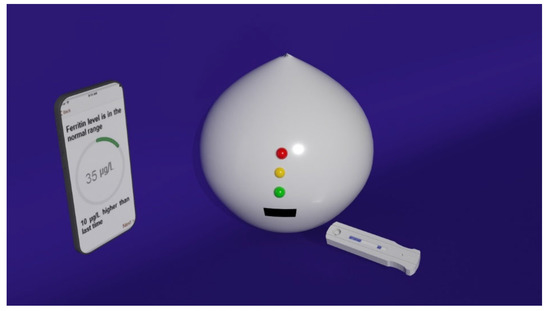
Figure 1.
Nutri
phone system overview illustrating the custom vitamin B12 test strip along with the app and the box reader.
Motivated by the need for a rapid and reliable diagnosis to counteract the 2014 outbreak of Ebola virus disease (EVD) in West Africa, in the work of Brangel [4][63], a POC test for Ebola virus detection was designed. In the developed test, the sample is applied onto the sample pad and migrates to the conjugation pad where complexes between the target analytes and antihuman antibody IgG–gold nanoparticle conjugates are formed. From the conjugation pad, then, the sample runs towards the analytical pad, where the targeted IgG serum antibodies against single or multiple recombinant Ebola viral proteins bind to detection lines, resulting in a visual red-purple line. In addition, a control line is introduced to validate the assay. Two platforms were developed for single detection by means of Sudan virus (SUDV) recombinant glycoprotein GP1–649 or multiple detection using the SUDV recombinant proteins VP40, NP and GP1–649. Over the course of time, five distinct species of the ebolavirus genus have been detected, four of which are etiological agents of the viral haemorrhagic Ebola virus disease (EVD); thus, the sensing platform has been upgraded for viral-subtype identification: in that case, a multiplexed detection line contains the recombinant glycoprotein (GP1−649) for different Ebola viral species such as SUDV, Ebola virus (EBOV) and Bundibugyo virus (BDBV). In addition, the sensing strip is combined with a compatible smartphone application for semi-quantitative detection. After inserting the patient data in the app login window, the analysis window opens: after aligning a red box between the test and control lines, the results are presented. Based on the relative intensity of the test line and on an evaluated cut-off threshold, the app determines if the result is positive or negative. In addition, by means of the app, data storage and sharing is possible, as well as geographical tagging of the tested individuals in Uganda.
2. Liquid Biopsy
The evaluation of the cancer mutational profile is crucial in cancer therapy, and in routine diagnostics is performed through a fragment of the primary tumour or metastasis; nevertheless, there are some disadvantages to this kind of test. For example, surgical intervention is required to obtain tumoral tissues, thus widely limiting the sampling, or the presence of multiple tumour sites leads to a complication in the cancer characterization. Additionally, the intra-tumour heterogeneity, especially in the case of a single biopsy test, could lead to untrustworthy results [5][64]. In this context, there is a strong need for analysis requiring non- or minimally invasive procedures through which real-time monitoring of the patient’s cancer molecular alterations is possible. Very recently, the analysis of circulating tumour cells (CTCs), taking the name of “liquid biopsy,” has created new avenues for cancer diagnostics, involving improved risk staging and evaluation, and early tumour detection, and, in cancer therapy, the control of tumour evolution as well as the early detection of relapse. Initially, a liquid biopsy was exploited to detect CTCs, cancer cells detached from a primary tumour and/or metastatic lesion(s) and traveling through the bloodstream to other parts of the body; however, now, it has been extended to other components released by the tumour in the body fluids, such as cell-free circulating nucleic acids (DNA, mRNA, and non-coding RNA such as micro-RNA or long non-coding RNA), “tumour-educated platelets” (TEPs) or vesicles such as exosomes [6][65]. Due to its characteristics, SERS has emerged as a new tool to perform liquid biopsies, with the final objective of developing fast and cheap diagnostics, and prognostic and predictive tools that can be employed in the point-of-care setting by simply scanning a biofluid sample with a laser [7][8][66,67]. A novel lateral flow strip assay for mutational analysis of ctDNA (cell-free circulating tumor DNA) in blood samples for liquid biopsy based on gold nanoparticles was developed in the work of Kalligosfyri [9][68]. The test strip is composed of an absorbent pad, a nitrocellulose membrane (diagnostic membrane) containing the test zone (TZ) and the control zone (CZ), a conjugate pad and an immersion pad. A plastic surface is used as a support for all the components, and the ends of the pads are overlapped so that the proper capillary flow of the running buffer through the strip is ensured. The CZ contains biotin molecules from b-BSA conjugates, capable of collecting the excess of AuNPs functionalized with streptavidin (SA-AuNPs). This zone guarantees that the lateral flow assay works properly and it must be always visible. The so-developed method involves ctDNA isolation, PCR amplification of the KRAS gene, and a multiplex primer extension (PEXT) reaction, so that the strip for the visual analysis is not yet completely ready as a POC device. Motivated by the significant death rate caused by the scarcity of sensitive analytical devices for the early detection of the disease and the lack of relevant biomarkers relevant to diagnosis, Tadimety et al., in their work [10][69], designed and developed a nanoplasmonic platform based on Au nanorods for the detection of pancreatic ductal adenocarcinoma, which has great potential for liquid biopsies. One significant biomarker in a liquid biopsy is the circulating tumour DNA (ctDNA) which, due to its very short half life in peripheral blood, is able to give information about genetic and epigenetic alterations occurring in the tumour. Due to a prevalence rate of 88% in pancreatic cancers, the mutations in the KRAS gene were selected as a target: the most common mutations are localized in coding base 35 of exon 2 and, between them, the c.35G > T mutation replacing glycine 12 with a residue of valine (G12V) was selected: due to its prevalence in 37% in pancreatic cancers and from a study [11][70], this is the most common mutation in the KRAS gene. Once the target of the test is selected, the nanoplasmonic platform is prepared: after activating the surface for the binding of the peptide nucleic acid (PNA) probe through NHS/EDC chemistry, the Au nanorods are later conjugated to the G12V mutation in the KRAS gene. To be specific, the structure of PNA is a hybrid between a nucleic acid and a protein, with the backbone of a protein and functional nitrogenous bases linked through a tertiary acetamide, and in order to realize how the selectivity towards the binding of the mutant over the wild type could be improved by introducing mutations into the peptide nucleic acid probe, a simulation of the DNA hybridization was carried out. From the simulation, a PNA with an additional mutation two base pairs away from the mutation of interest showed the best selectivity between mutant and wild-type capture, so it was selected to functionalize the Au nanorods’ surface and the hybridization with the DNA was performed using UV-spectroscopy. As a consequence of hybridization, the LSPR peak in the spectra shifted towards longer wavelengths, linearly depending on ctDNA concentrations, so that, using solutions at different concentrations, it was possible to calculate an LOD of 2 nanograms of ctDNA per milliliter. The sensing transducer was tested in both buffer and spiked healthy-patient serum, assuring a sequence-specific capture of ctDNA and a strategy to improve the selectivity towards the requested point mutations by means of the accurate design of the probe. The high specificity displayed in the results lays the groundwork for realizing multiplexing and arrayed sensor designs by integrating the principles outlined in the work on a chip.
3. Wearable Plasmonic Point-of-Care Devices
3. Wearable Plasmonic POC Devices
In light of personalized healthcare, wearable biosensors have attracted growing interest as a new real-time and non-invasive detection technology. To obtain a deep insight into the human body at a more profound molecular level, several kinds of wearable biosensors are based on plasmonic nanoparticles. The work of Mogera [12][71] describes a wearable plasmonic paper-based microfluidic device for continuous analysis of sweat. Most of the sensing systems exploit enzymes and antibodies, but they tend to degrade over time, thus undermining their performances and, in order to create a more stable device for continuous measurement, a microfluid device integrated with plasmonic paper was designed and then tested for the detection of uric acid (UA), a risk biomarker for cardiovascular diseases, kidney diseases, and type-2 diabetes. The sensing device has a layered structure: at the bottom is placed a stretchable medical-grade double-sided adhesive tape, forming a mechanically robust interface between the skin and the adjacent layer of the device. A black carbon double-sided adhesive, blocking laser radiation to avoid skin damage during in-situ SERS analysis, is enclosed between the double-sided adhesive and the adjacent layer, the microfluidic system. In the developed device, the microfluidic system is an effective microfluidic channel made of cellulose chromatography paper with a serpentine design, providing flexibility and stretchability to accommodate skin deformation without altering device performance, which transports the excreted sweat. A large inlet opening is able to enhance the access to sweat glands, thus maximizing the collection of sweat, whereas at the end of the microchannel is localized an outlet to avoid back pressure. The layer above the microfluidic channel is the plasmonic sensor. It consists of plasmonic paper immobilized in different locations along the microfluidic channel and it is the element of the device that quantifies the concentration of analytes in sweat by means of SERS spectroscopy. The plasmonic paper is formed by gold nanorods (AuNRs) synthesized according to a seed-mediated growth protocol adsorbed on a chromatography-grade filter paper and cut into squares of 2 mm × 2 mm or circles with a 1 mm diameter. All the layers are then encapsulated with a polydimethylsiloxane (PDMS) film: it displays well-defined Raman bands serving as a reference to quantify the analytes of interest in the sweat and, in addition, it is optically transparent, it prevents contamination from the environment, and it minimizes the evaporation of sweat. After testing the mechanical properties, the flow characteristics, and the SERS performance of the sensing layer, the device was exploited to detect UA in healthy human subjects. By means of a portable Raman spectrometer with a flexible fibre probe focused on the sensing part of the system, an SERS spectrum of the sweat was collected and the concentration of UA quantified. For this wearable plasmonic paper-based microfluidic device, an LOD of 1 μM was revealed; however, despite its great potential as a POC device, it needs still some improvements. In fact, the readout is based on a handheld Raman spectrometer, it was not developed as an integrated electronic readout system (Figure 2).
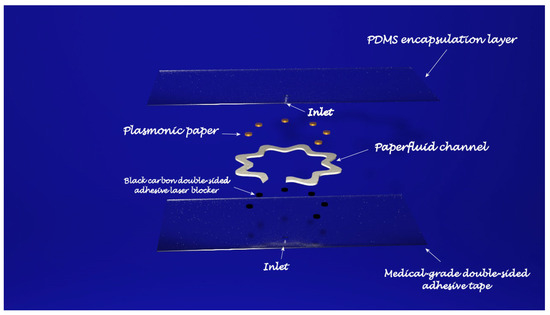
In the work of Koh [13][72], a wearable SERS sensor was developed as a skin-adhesive patch and successively tested to detect 2-fluoromethamphetamine (2-FMA), which is an analogue of methamphetamine. The basement layer is composed of a silk fibroin protein film (SFF): it contains nanopores which were created in the β-sheets domains of the SFF matrix and are able to filter the molecules. Anyway, the size of the nanopores in the SFF layer can be controlled during the successive process of crystallization, thus allowing a different grade of permeability depending on the nanopore size. A layer of silver nanowires (AgNWs) was then deposited onto the SFF film: inside the layer, the AgNWs form a network with a random structure and at the intersections, hotspots are created with a high density. The SFF/AgNWs were then transferred to an adhesive, transparent dermal patch which protects the wearable sensor from contamination and oxidation when it is worn. In molecular penetration tests, about 90 min are required for the molecules to diffuse through the SFF layer until the AgNWs. Once assembled, the SERS patch was tested for analysis using different water-soluble Raman-reported dyes (4-aminothiophenyl, DAPI, methylene blue, rhodamine B, eosin B, eosin Y and erythrosin B) in order to determine the molecular weight beyond which the SERS performances are reduced; the results showed that the system is well suited for Raman reporters with a molecular weight lower than eosin B. molecules, as higher molecular weights were not filtered by the SFF layer. To evaluate the performances of the wearable SERS patch, 2-FMA was selected. In this case, a solution of 2-FMA was added to simulate human sweat, and the obtained solution was then dropped like sweat on human-cadaver skin. The human-cadaver skin was then covered with the sensing patch and, after two hours, the SERS spectra were measured. Despite the good potential as a POC device and the ability to accumulate molecules in the SFF layer, so that the analysis can be performed long after the molecule has been released in the sweat, the wearable SERS patch cannot be used for real-time monitoring, because analytes molecules took about 1,5 h to reach the AgNWs layer, so that the analysis is not very fast.
4. Multiplexed Analysis with Surface-Enhanced Raman Spectroscopy Nanotags
3. Multiplexed Analysis with SERS Nanotags
Acute myocardial infarction (AMI) is recognized as one of the most immediately life-threatening problems and it is among the main causes of death in the world; for its quick diagnosis, the simultaneous detection of cardiac biomarkers such as cardiac troponin I (cTnI), creatine kinase-MB isoenzymes (CK-MB) and myoglobin (Myo) is required. On this basis, lives could be more easily saved by elevating the level in AMI tracking through the multiplex and quantitative detection of these three biomarkers performed with a rapid LFA strip test. Taking into account the increasing interest in the production of SERS nanotags for rapid and accurate analysis, Zhang, in his work [14][73], developed an LFA strip for the multiplexed and quantitative detection of the three cardiac biomarkers for early AMI diagnosis integrating a specifically created SERS nanotag. The SERS LFA strip is formed by five components: a backing pad, a sample pad, a conjugate pad, a nitrocellulose (NC) membrane and an adsorbing pad. To the lower part of the backing pad are fixed the sample and the conjugate pad; the NC membrane is fixed to the middle part of the backing pad whereas to the higher part is fixed the adsorbing pad. To provide a continuous stream of the developing solution from the sample to the adsorption pad by means of capillary forces, the components are overlapped. In the conjugate pad are the Myo, cTnI, and CK-MB detection antibody conjugated with the SERS nanotags. In place of gold colloid, silver–gold core-shell bimetallic nanotags encapsulating Nile blue A (NBA) molecules (the Raman reports) in the interior gap between the two metals (AgNBA@Au) were synthesised as SERS nanotags. Onto the NC membrane were placed the three test lines and the control line: the test lines were prepared by separately spotting to capture the antibody against CK-MB, the capture antibody against Myo and the capture antibody against cTnI, while the control line was produced with goat anti-mouse IgG. To collect the SERS spectra coming from the SERS nanotags conjugated with the analyte in the test line, a linear dynamic range (LDR) of 0.01–500 ng mL−1, 0.01–50 ng mL−1, and 0.02–90 ng mL−1 is needed for Myo, cTnI, and CK-MB. With results comparable with chemiluminescence immunoassay (CLIA) and its ease of use arising from pre-treatments not being required, the sensitive SERS LFA strips hold great potential for point-of-care testing technologies (POCT) for AMI diagnosis with applications in health monitoring and clinical alerts. In a later work [15][74], the authors developed a test strip with a single test line comprising the capture antibodies against the three cardiac biomarkers CK-MB, cTnI, and Myo with the aim to improve the performances of the devices by reducing the analysis time. In Table 1 a comparison of the considered POC devices has been reported.
Table 1.
Comparison of the advantages and limitations of the considered POC devices.
| System | Analyte | Advantages | Limitations | Reference | ||||
|---|---|---|---|---|---|---|---|---|
| Colorimetric sensing of urea based on the generation of silver nanoparticles performed through image analysis with a smartphone. | Urea. | -High selectivity toward various interferents in human urine. -High sensitivity. -Wide detection range. -No need for complicated instrumentation. -Real-time responses. |
- | [1] | [60] | |||
| Iron | phone system. | Ferritin. | -High sensitivity. -Real-time response. -No need for bulky equipment. |
Ready as POC device. | [2] | [61] | ||
| Nutri | phone system. | Vitamin B | 12 | . | -High sensitivity. -Real-time response. -No need for bulky equipment. |
Ready as POC device. | [3] | [62] |
| Smartphone lateral flow point-of-care test for Ebola virus. | Ebola virus. | -High sensitivity and specificity. -Rapid and portable analysis. -Data storage and sharing as well as geographical tagging of the tested individuals. |
Further work to evaluate sensitivity and specificity in larger cohort groups, need for studies on assay compatibility under a broader range of environmental conditions at POC and vaccine evaluation. | [4] | [63] | |||
| Lateral flow assays based on AuNPs. | -Normal KRAS gene and three of the most significant/common mutations in the KRAS gene in cell-free DNA. |
-High detectability. -Good specificity. -Good reproducibility. -Visual detection. -Low cost. -Rapid analysis time. |
The whole protocol includes techniques such as DNA isolation, polymerase chain reaction (PCR) and Primer extension reaction (PEXT), requiring trained personnel. | [9] | [68] | |||
| Gold nanorods in solution. | -KRAS gene associated with pancreatic ductal adenocarcinoma in circulating tumor DNA (ctDNA). | -High sensitivity. | -Need for bulky instrumentation such as UV-spectrometer. | [10] | [69] | |||
| Wearable plasmonic paper-based microfluidic device for continuous analysis of sweat. | -Uric acid (UA) in sweat at physiological and pathological concentrations. | -Continuous, real-time, label-free analysis of biomolecules. -Performances not compromised by the poor stability of biorecognition elements -High sensitivity -High specificity |
-Need for an optimized miniaturized Raman spectrometer as read-out system. | [12] | [71] | |||
| SERS patch sensor based on plasmonic silver nanowire (AgNW). | 2-fluoromethamphetamine (2-FMA). | -Excellent skin biocompatibility. -Flexibility. -Durability for use as a wearable sensor. -High sensitivity. -High specificity. -Direct label-free detection. |
-Need for a miniaturized Raman spectrometer. -Quasi real-time monitoring: necessity to wait more than 90 min to start analysis. |
[13] | [72] | |||
| Core-shell SERS nanotag-based multiplex LFA (SERS LFA) for rapid and quantitative detection of three cardiac biomarkers. | -cardiac troponin I (cTnI). -creatine kinase-MB isoenzymes (CK-MB). -myoglobin (Myo). |
-Ultrasensitive. -Fast. -Low cost. -Easy to use. -Free of sample pretreatment and professional labors. |
-Need for a miniaturized and portable Raman spectrometer, but they still must be improved. | [14] | [73] | |||
| Core-shell SERS nanotag-based multiplex LFA (SERS LFA) for rapid and quantitative detection of three cardiac biomarkers. | -cardiac troponin I (cTnI). -creatine kinase-MB isoenzymes (CK-MB). -myoglobin (Myo). |
-Ultrasensitive. -Faster quantification of three cardiac biomarkers. -Low cost. -Easy to use. -Free of sample pretreatment and professional labor. |
-Need for a miniaturized and portable Raman spectrometer, but they still must be improved. | [15] | [74] |
