Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Dean Liu and Version 1 by Koushik Halder.
RNA interference (RNAi) has inarguably been a revolutionary discovery in the field of biology in the last several decades. The discovery of small (20–30 nucleotide long) non-coding RNAs that can regulate genes and the genome completely transformed RNA biology. These small RNAs can guide effector proteins targeting any complementary nucleotide sequence through the RNAi pathway, thereby downregulating its expression level. Napoli and Jorgensen first reported small RNA-mediated gene regulation in plants while working with chalcone synthase (CHS) in petunia.
- small non-coding RNAs
- siRNA
- miRNA
- tasiRNA
1. Biogenesis of Short Interfering RNAs
dsRNA-mediated post-transcriptional gene silencing (PTGS) symbolizes the cellular defense mechanism protecting it from foreign nucleic acids of invading viruses or transposons [18,19][1][2]. Such integration of alien genes produces dsRNAs, which act as a guide for sequence-specific RNA degradation and are believed to be associated with maintaining the silencing process for a long time [20,21][3][4]. The processing of these long dsRNAs into 21–24 nt long short-interfering RNAs (siRNAs) is facilitated by Dicer, a ribonuclease III enzyme [22][5] (Figure 1A). The presence of these siRNAs was first reported by Hamilton and Baulcombe in plant tissues that showed virus-induced PTGS [23][6]. These siRNAs were later found to be present also in Drosophila melanogaster embryo lysate [24][7], where the added synthetic 20–22 nt RNA duplexes can efficiently target and cleave mRNAs at 21 nt intervals [25][8]. That is why these 21 nt long RNAs were called siRNAs or silencing RNAs. Dicer-mediated processing is coupled with other co-factors, and these siRNAs are finally loaded onto the RNA-induced silencing complex (RISC), a member of the argonaute protein family [26][9]. During incorporation within the RISC, one strand called the “passenger strand” is dissociated by the activity of AGO2, which is encoded by the gene AGO2 (argonaute RISC catalytic component 2), whereas the other “guide strand” that serves as a guide for RNA-directed sequence-specific silencing stays within the complex, forming the mature RISC [27,28][10][11]. Targeted mRNA with sequence complementarity with the 21 nt long guide-siRNA within the mature RISC is cleaved between the 10th and 11th nucleotide (from the 5′ end) by the PIWI domain of the AGO2 protein, generating products containing 5′-monophosphate and 3′-hydroxyl termini [29,30][12][13]. These cleaved products are rapidly degraded by the endogenous exonuclease activity due to the lack of 5′ capping or a 3′poly(A) tail [31][14]. Apart from the above-mentioned RNAi, plants [20,32][3][15], fungi [33][16], and C. elegans [21,34][4][17] possess a special enzyme called RNA-dependent RNA polymerases (RdRPs), which can produce additional dsRNAs for amplifying the RNAi response. Such dsRNAs are synthesized in a primer-independent manner using the targeted mRNA as a template. It is subsequently processed by Dicer to produce more siRNAs, thereby facilitating the recycling of the RISC complex [35][18] (Figure 1B).
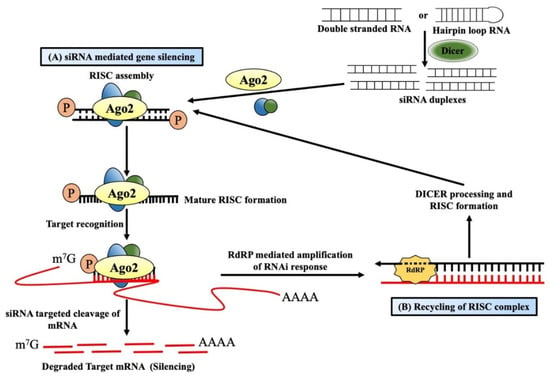
Figure 1. Mechanism of short-interfering RNA-mediated gene silencing. (A) Long double-stranded RNAs/hairpin loop RNAs from alien genes are processed into short interfering RNAs (siRNAs) by the Dicer/TRBP (the human immunodeficiency virus transactivating response RNA binding domain) complex and finally become incorporated into RNA-induced silencing complex (RISC). One strand (passenger strand) is degraded from the RNA duplex, and the other strand (guide strand), along with argonaute 2, forms the active RISC. The guide strand guides the active RISC to target and cleaves the complementary mRNAs into the cytosol, resulting in gene silencing. (B) RNAi response in plants and worms generally becomes amplified by the RNA-dependent RNA polymerase enzymes (RdRPs). RdRPs and RISC use targeted mRNAs as a template to generate double-stranded RNAs dsRNAs, which then are processed by the Dicer into secondary siRNAs. These siRNAs eventually amplify the RNAi effect in the system.
2. RNA-Induced Silencing Complex: The Versatile Gene Silencing Complex
Although there are diverse ways to regulate gene expressions using RISC, two significant incidents are common for all types. Firstly, every RISC should comprise one argonaute protein family member, and secondly, at its core, a small RNA should guide the RISC to target mRNAs through Watson–Crick base pairing [36][19]. Each class of small RNAs in eukaryotes, such as siRNA/miRNA/piRNA (only in animals), together with the AGO protein family, forms the ribonucleoprotein RISC. Our current understanding of RISC (from birth to death) has been summarized here.
The Argonaute family of proteins is at the heart of RISC-mediated gene regulation [37,38][20][21]. There are four functional domains in AGO proteins: PIWI-AGO-Zwille (PAZ), Middle (MID), N-terminal (N), and PIWI (Figure 2) [37,39][20][22]. There are two linkers (L1, L2), out of which L1 connects the N and PAZ domains, whereas L2 supports the N-L1-PAZ structure connecting it to the MID-PIWI lobe [40,41][23][24]. The PAZ domain of AGO proteins contains a pocket that interacts with and binds the guide strand from the 3′ end [42][25]. A conserved sequence of a catalytic tetrad (Asp-Glu-Asp-His/Asp) can be found in the PIWI domain of some AGO proteins that are responsible for the target mRNA degradation. This catalytic domain is also responsible for the cleavage of some passenger strands before it gets ejected [39][22]. In the initial phase of RISC assembly, a small RNA duplex is loaded onto an empty AGO protein with the help of Hsp70/Hsp90 chaperons and forms the pre-RISC [43][26]. One of the strands, which is less stable and likely to be adenine/uridine (AU) rich, is preferably able to be the guide strand. This type of strand selection is asymmetric and generally depends on the difference in the thermodynamic stability of two ends of the RNA duplex [44,45][27][28]. At the initial stage of passenger strand separation, the N domain disrupts the 3′ end base-pairing of the guide strand to help open the RNA duplex [46][29]. Then, the passenger strand is sliced at a position opposite the guide strand’s 10th and 11th nucleotide (g10 and g11) by the catalytic activity of the AGO protein [27,28,47][10][11][30]. After passenger strand separation, the mature RISC binds to the targeted mRNA guided by the guide strand and either slices it directly or induces translational repression by recruiting necessary proteins [48,49][31][32]. Although AGO and small RNAs are short-lived, once the RISC is formed, those two tend to be long-lived [50,51][33][34]. Generally blank/unloaded AGO proteins are degraded by the autophagy pathway [52[35][36],53], whereas the target RNA-directed miRNA degradation (TDMD) targeted RISC are degraded by the ubiquitin–protease system [54,55][37][38].
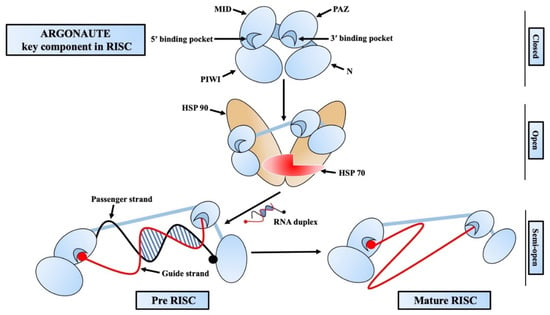
Figure 2. Mechanism of RNA-induced silencing complex (RISC) assembly. Argonaute has four functional domains: PIWI-AGO-Zwille (PAZ), Middle (MID), N-terminal (N), and PIWI (Piwi/Argonaute/Zwille). An empty argonaute loads an RNA duplex with the help of HSP70/HSP90 and forms the pre-RISC. The passenger strand (black strand) is ejected from the pre-RISC, and the guide strand (red strand) and argonaute form the mature/active RISC that start targeting the complimentary mRNAs to assert gene silencing.
3. Short Interfering RNA Mediated Silencing in Plants
The siRNA-mediated gene silencing mechanism has been exploited extensively over the years for crop protection against biotic stresses and as a platform for overall crop improvement. RNAi-mediated silencing requires the design of a hairpin loop structure containing both the sense and anti-sense strand of the targeted gene separated by an intron sequence. Such a construct will create hpRNAs in plants that are cleaved by Dicer and form siRNAs, which trigger the RNAi pathway to silence the targeted gene [56][39]. Pests such as insects, nematodes, viruses, and bacteria pose a severe threat to agricultural produce. RNAi has been used to make pest-resistant crops by targeting essential genes in the pests and silencing them by host plant-mediated RNAi (host-induced gene silencing—HIGS) [57][40].
The two-spotted spider mite is one of the deadliest plant pests; it can attack more than three thousand crops and feeds mainly on Chinese cabbage. RNAi-mediated targeting against the COPB2 gene of this pest resulted in an almost 100% mortality rate [58][41]. Similarly, transgenic potato plants expressing the molting-associated EcR gene showed enhanced resistance against the deadly Colorado potato beetle [59][42]. The root-knot nematode Meloidogyne incognita causes colossal damage to agricultural products worldwide. RNAi-mediated simultaneous silencing of Mi-flp1, Mi-flp12, and Mi-flp18 genes resulted in enhanced resistance against this nematode [60][43]. An extensive list of such siRNA-mediated gene silencing to improve desirable traits in plants is listed in Table 2. Because RNAi-derived plants have been categorized as genetically modified crops (GM crops) and have been the subject of bitter controversy, the exogenous application of RNAi-inducing dsRNA-based bio-pesticides is gaining popularity, as it provides a non-transgenic approach [61][44]. Tenllado et al. first reported the effective foliar application of dsRNAs targeting the Alfalfa mosaic virus (AMV), Tobacco etch virus (TEV), and Pepper mild mottle virus (PMMoV) in 2001 [62][45]. However, the authors did mention that the commercial success of such topical application of dsRNAs will depend on two critical parameters: cost-effectiveness and an optimized mode of delivery. In recent years, several studies have been conducted to deliver such topical solutions of dsRNAs with impeccable efficiency and target specificity [62,63,64][45][46][47]. Topically applied dsRNA-based biopesticides have high species specificity, low levels of toxicity, and a minimal environmental effect compared to traditional pesticides. If their distribution and usage can be regulated in a precautionary way, dsRNA-based biopesticides can revolutionize the integrated pest-management system [65][48].
4. Tweaking the siRNAs to Improve Desirable Traits in Plants
siRNAs are well known for their silencing role in the case of viral RNAs. They play a master role in regulating plant defense machinery against potential pathogens such as bacteria, viruses, fungi, oomycetes, and other parasitic plants. Recent research has provided evidence of the ability of siRNAs to suppress fungus and oomycetes by silencing specific pathogen genes related to pathogenesis. Thus, scientists have precisely concluded that siRNAs are a potential concoction of diverse gene sequences and are used as a “shotgun” which targets random genes of the pathogen with great efficiency [66][49]. After discovering the antiviral factor, virus-derived siRNAs (vsiRNAs) in tobacco infected with potato virus, there has been a breakthrough in plant immunity. These viral dsRNAs are directly targeted by plant DCLs, resulting in 21–24 nt long primary vsiRNAs. It has been established that these 21 nt long vsiRNAs specifically silence the detrimental viral RNAs through Post Transcriptional Gene Silencing (PTGS) [67][50]. The functions of vsiRNAs can also be categorized into two parts. One part is when the vsiRNAs degrade the intruder viral genome and render antiviral tolerance to plants. On the other side, some vsiRNAs silence host gene expression and manipulate host resistance towards viral attack. During tomato yellow leaf curl virus (TYLCV) infection, it has been observed that vsiRNAs are utilized by TYLCV to silence SILNR1, a long noncoding RNA (lncRNA) associated with antiviral defense. vsiRNAs obtained from wheat yellow mosaic virus (WYMV) down-regulate host genes and activated broad-spectrum host immunity [68][51]. Another category of siRNA, which was first seen in Arabidopsis, which also participates in antiviral defense, is virus-activated siRNA (vasiRNA) [69][52]. In the case of antiviral defense, the vsiRNAs are generated from the viral genome, which safeguards the plants by destroying the viral RNA. However, in the case of non-viral plant pathogens, endogenous siRNA-orchestrated gene silencing is instantly triggered to alter the gene expression associated with plant immunity [66][49]. The components of the siRNA pathway interact among themselves and others to orchestrate plant immunity. RDR6 is essential for plant immunity because it aids in producing secondary siRNAs and silencing signals. For example, in rice, shl2-rol, which is a mutant line of the rice gene OsRDR6 results in severe infection symptoms when the plant is attacked by Xanthomonas oryzae PV. oryzae, thus proving the importance of RDR6-dependent siRNAs in rendering tolerance against bacteria [70][53].
Artificial siRNAs are produced in plants via Host-Induced Gene Silencing (HIGS) to silence the deadly pathogen genes causing infection. A well-tested example is transgenic barley and wheat, which express artificial siRNAs that target gene Avra10 and display increased resistance to Blumeria graminis, the causative agent of powdery mildew disease [71][54]. HIGS via engineered dsRNA resists parasitic plants. This strategy has been demonstrated in transgenic tobacco that expresses dsRNA against transcription factors controlling haustoria development and reduced vigor in Cuscuta patagonia. Similarly, Orobanche aegyptiaca has been recorded to grow and feed on tomatoes. The above examples highlight host-produced synthetic siRNAs’ significant role in controlling parasitic plants.
References
- Fire, A. RNA-Triggered Gene Silencing. Trends Genet. 1999, 15, 358–363.
- Carthew, R.W. Gene Silencing by Double-Stranded RNA. Curr. Opin. Cell Biol. 2001, 13, 244–248.
- Dalmay, T.; Hamilton, A.; Rudd, S.; Angell, S.; Baulcombe, D.C. An RNA-Dependent RNA Polymerase Gene in Arabidopsis Is Required for Posttranscriptional Gene Silencing Mediated by a Transgene but Not by a Virus. Cell 2000, 101, 543–553.
- Smardon, A.; Spoerke, J.M.; Stacey, S.C.; Klein, M.E.; MacKin, N.; Maine, E.M. EGO-1 Is Related to RNA-Directed RNA Polymerase and Functions in Germ-Line Development and RNA Interference in C. Elegans. Curr. Biol. 2000, 10, 169–178.
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366.
- Hamilton, A.J.; Baulcombe, D.C. A Species of Small Antisense RNA in Posttranscriptional Gene Silencing in Plants. Science 1999, 286, 950–952.
- Zamore, P.D.; Tuschl, T.; Sharp, P.A.; Bartel, D.P. RNAi: Double-Stranded RNA Directs the ATP-Dependent Cleavage of MRNA at 21 to 23 Nucleotide Intervals. Cell 2000, 101, 25–33.
- Elbashir, S.M.; Lendeckel, W.; Tuschl, T. RNA Interference Is Mediated by 21- and 22-Nucleotide RNAs. Genes Dev. 2001, 15, 188–200.
- Hammond, S.M.; Boettcher, S.; Caudy, A.A.; Kobayashi, R.; Hannon, G.J. Argonaute2, a Link between Genetic and Biochemical Analyses of RNAi. Science 2001, 293, 1146–1150.
- Matranga, C.; Tomari, Y.; Shin, C.; Bartel, D.P.; Zamore, P.D. Passenger-Strand Cleavage Facilitates Assembly of SiRNA into Ago2-Containing RNAi Enzyme Complexes. Cell 2005, 123, 607–620.
- Rand, T.A.; Petersen, S.; Du, F.; Wang, X. Argonaute2 Cleaves the Anti-Guide Strand of SiRNA during RISC Activation. Cell 2005, 123, 621–629.
- Tomari, Y.; Zamore, P.D. Perspective: Machines for RNAi. Genes Dev. 2005, 19, 517–529.
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21±nucleotide RNAs Mediate RNA Interference in Cultured Mammalian Cells Sayda. Nature 2001, 411, 494–498.
- Elbashir, S.M.; Martinez, J.; Patkaniowska, A.; Lendeckel, W.; Tuschl, T. Functional Anatomy of SiRNAs for Mediating Efficient RNAi in Drosophila Melanogaster Embryo Lysate. EMBO J. 2001, 20, 6877–6888.
- Mourrain, P.; Béclin, C.; Elmayan, T.; Feuerbach, F.; Godon, C.; Morel, J.B.; Jouette, D.; Lacombe, A.M.; Nikic, S.; Picault, N.; et al. Arabidopsis SGS2 and SGS3 Genes Are Required for Posttranscriptional Gene Silencing and Natural Virus Resistance. Cell 2000, 101, 533–542.
- Cogoni, C.; Macino, G. Gene Silencing in Neurospora Crassa Requires a Protein Homologous to RNA-Dependent RNA Polymerase. Nature 1999, 399, 166–169.
- Brown, R.D.; Mattoccia, E.; Tocchini-Valentini, G.P. On the Role of RNA in Gene Amplification. Acta Endocrinol. Suppl. 1972, 168, 307–318.
- Mello, C.C.; Conte, J.D. Revealing the World of RNA Interference. Nature 2004, 431, 338–342.
- Pratt, A.J.; MacRae, I.J. The RNA-Induced Silencing Complex: A Versatile Gene-Silencing Machine. J. Biol. Chem. 2009, 284, 17897–17901.
- Peters, L.; Meister, G. Argonaute Proteins: Mediators of RNA Silencing. Mol. Cell 2007, 26, 611–623.
- Meister, G. Argonaute Proteins: Functional Insights and Emerging Roles. Nat. Rev. Genet 2013, 14, 447–459.
- Nakanishi, K.; Weinberg, D.E.; Bartel, D.P.; Patel, D.J. Structure of Yeast Argonaute with Guide RNA. Nature 2012, 486, 368–374.
- Elkayam, E.; Kuhn, C.D.; Tocilj, A.; Haase, A.D.; Greene, E.M.; Hannon, G.J.; Joshua-Tor, L. The Structure of Human Argonaute-2 in Complex with MiR-20a. Cell 2012, 150, 100–110.
- Wang, Y.; Sheng, G.; Juranek, S.; Tuschl, T.; Patel, D.J. Structure of the Guide-Strand-Containing Argonaute Silencing Complex. Nature 2008, 456, 209–213.
- Parker, J.S.; Roe, S.M.; Barford, D. Structural Insights into MRNA Recognition from a PIWI Domain-SiRNA Guide Complex. Nature 2005, 434, 663–666.
- Iwasaki, S.; Kobayashi, M.; Yoda, M.; Sakaguchi, Y.; Katsuma, S.; Suzuki, T.; Tomari, Y. Hsc70/Hsp90 Chaperone Machinery Mediates ATP-Dependent RISC Loading of Small RNA Duplexes. Mol. Cell 2010, 39, 292–299.
- Fomenko, A.I.; Donchenko, G.V.; Stepanenko, S.P. Effect of Withdrawal of Phenazepam and Nicotinamide on the State of the Systems of Reception of Benzodiazepines and NAD. Ukr Biokhim Zh 1996, 68, 24–25.
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional SiRNAs and MiRNAs Exhibit Strand Bias. Cell 2003, 115, 209–216.
- Wang, Y.; Juranek, S.; Li, H.; Sheng, G.; Wardle, G.S.; Tuschl, T.; Patel, D.J. Nucleation, Propagation and Cleavage of Target RNAs in Ago Silencing Complexes. Nature 2009, 461, 754–761.
- Miyoshi, K.; Tsukumo, H.; Nagami, T.; Siomi, H.; Siomi, M.C. Slicer Function of Drosophila Argonautes and Its Involvement in RISC Formation. Genes Dev. 2005, 19, 2837–2848.
- Djuranovic, S.; Nahvi, A.; Green, R. MiRNA-Mediated Gene Silencing by Translational Repression Followed by MRNA Deadenylation and Decay. Science 2012, 336, 237–240.
- Iwakawa, H.O.; Tomari, Y. Life of RISC: Formation, Action, and Degradation of RNA-Induced Silencing Complex. Mol. Cell 2022, 82, 30–43.
- Winter, J.; Diederichs, S. Argonaute Proteins Regulate MicroRNA Stability: Increased MicroRNA Abundance by Argonaute Proteins Is Due to MicroRNA Stabilization. RNA Biol. 2011, 8, 1149–1157.
- Vaucheret, H.; Vazquez, F.; Crété, P.; Bartel, D.P. The Action of ARGONAUTE1 in the MiRNA Pathway and Its Regulation by the MiRNA Pathway Are Crucial for Plant Development. Genes Dev. 2004, 18, 1187–1197.
- Derrien, B.; Baumberger, N.; Schepetilnikov, M.; Viotti, C.; De Cillia, J.; Ziegler-Graff, V.; Isono, E.; Schumacher, K.; Genschik, P. Degradation of the Antiviral Component ARGONAUTE1 by the Autophagy Pathway. Proc. Natl. Acad. Sci. USA 2012, 109, 15942–15946.
- Kobayashi, H.; Shoji, K.; Kiyokawa, K.; Negishi, L.; Tomari, Y. VCP Machinery Mediates Autophagic Degradation of Empty Argonaute. Cell Rep. 2019, 28, 1144–1153.e4.
- Shi, C.Y.; Kingston, E.R.; Kleaveland, B.; Lin, D.H.; Stubna, M.W.; Bartel, D.P. The ZSWIM8 Ubiquitin Ligase Mediates Target-Directed MicroRNA Degradation. Science 2020, 370, eabc9359.
- Han, J.; Lavigne, C.A.; Jones, B.T.; Zhang, H.; Gillett, F.; Mendell, J.T. A Ubiquitin Ligase Mediates Target-Directed MicroRNA Decay Independently of Tailing and Trimming. Science 2020, 370, eabc9546.
- Rajam, M.V. RNA Silencing Technology: A Boon for Crop Improvement. J. Biosci. 2020, 45, 118.
- Ghag, S.B. Host Induced Gene Silencing, an Emerging Science to Engineer Crop Resistance against Harmful Plant Pathogens. Physiol. Mol. Plant Pathol. 2017, 100, 242–254.
- Shin, Y.H.; Lee, S.H.; Park, Y.D. Development of Mite (Tetranychus Urticae)-Resistant Transgenic Chinese Cabbage Using Plant-Mediated RNA Interference. Hortic. Environ. Biotechnol. 2020, 61, 305–315.
- Hussain, T.; Aksoy, E.; Çalışkan, M.E.; Bakhsh, A. Transgenic Potato Lines Expressing Hairpin RNAi Construct of Molting-Associated EcR Gene Exhibit Enhanced Resistance against Colorado Potato Beetle (Leptinotarsa Decemlineata, Say). Transgenic. Res. 2019, 28, 151–164.
- Banakar, P.; Hada, A.; Papolu, P.K.; Rao, U. Simultaneous RNAi Knockdown of Three FMRFamide-like Peptide Genes, Mi-Flp1, Mi-Flp12, and Mi-Flp18 Provides Resistance to Root-Knot Nematode, Meloidogyne incognita. Front. Microbiol. 2020, 11, 573916.
- Halder, K.; Chaudhuri, A.; Abdin, M.Z.; Majee, M.; Datta, A. RNA Interference for Improving Disease Resistance in Plants and Its Relevance in This Clustered Regularly Interspaced Short Palindromic Repeats-Dominated Era in Terms of DsRNA-Based Biopesticides. Front. Plant Sci. 2022, 13, 885128.
- Tenllado, F.; Díaz-Ruíz, J.R. Double-Stranded RNA-Mediated Interference with Plant Virus Infection. J. Virol. 2001, 75, 12288–12297.
- McLoughlin, A.G.; Wytinck, N.; Walker, P.L.; Girard, I.J.; Rashid, K.Y.; De Kievit, T.; Fernando, W.G.D.; Whyard, S.; Belmonte, M.F. Identification and Application of Exogenous DsRNA Confers Plant Protection against Sclerotinia Sclerotiorum and Botrytis Cinerea. Sci. Rep. 2018, 8, 7320.
- Mitter, N.; Worrall, E.A.; Robinson, K.E.; Xu, Z.P.; Carroll, B.J. Induction of Virus Resistance by Exogenous Application of Double-Stranded RNA. Curr. Opin. Virol. 2017, 26, 49–55.
- Höfle, L.; Biedenkopf, D.; Werner, B.T.; Shrestha, A.; Jelonek, L.; Koch, A. Study on the Efficiency of DsRNAs with Increasing Length in RNA-Based Silencing of the Fusarium CYP51 Genes. RNA Biol. 2020, 17, 463–473.
- Fletcher, S.J.; Reeves, P.T.; Hoang, B.T.; Mitter, N. A Perspective on RNAi-Based Biopesticides. Front. Plant Sci. 2020, 11, 51.
- Kong, X.; Yang, M.; Le, B.H.; He, W.; Hou, Y. The Master Role of SiRNAs in Plant Immunity. Mol. Plant Pathol. 2022, 23, 1565–1574.
- Wang, X.B.; Wu, Q.; Ito, T.; Cillo, F.; Li, W.X.; Chen, X.; Yu, J.L.; Ding, S.W. RNAi-Mediated Viral Immunity Requires Amplification of Virus-Derived SiRNAs in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 484–489.
- Liu, P.; Zhang, X.; Zhang, F.; Xu, M.; Ye, Z.; Wang, K.; Liu, S.; Han, X.; Cheng, Y.; Zhong, K.; et al. A Virus-Derived SiRNA Activates Plant Immunity by Interfering with ROS Scavenging. Mol. Plant 2021, 14, 1088–1103.
- Cao, M.; Du, P.; Wang, X.; Yu, Y.Q.; Qiu, Y.H.; Li, W.; Gal-On, A.; Zhou, C.; Li, Y.; Ding, S.W. Virus Infection Triggers Widespread Silencing of Host Genes by a Distinct Class of Endogenous SiRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14613–14618.
- Wagh, S.G.; Alam, M.M.; Kobayashi, K.; Yaeno, T.; Yamaoka, N.; Toriba, T.; Hirano, H.Y.; Nishiguchi, M. Analysis of Rice RNA-Dependent RNA Polymerase 6 (OsRDR6) Gene in Response to Viral, Bacterial and Fungal Pathogens. J. Gen. Plant Pathol. 2016, 82, 12–17.
More
