Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Sirius Huang and Version 1 by Sony Kumari.
Citrus, belonging to the Rutaceae family, is a commercial fruit worldwide, and it is mainly recognized for its nutritional, anti-oxidant, and significant medicinal properties. Citruses are a group of multifaceted fruit crops with a rich traditional knowledge, deeply rooted in ethnic culture, and the fruits have been considered to be health-protecting and health-promoting food supplements since ancient times.
- citrus
- origin
- biodiversity
- production
1. Citrus Origin and Domestication
The Citrus term is originated from ‘Kedros’, a Greek term which denotes trees such as cidar, pine, and cypress. Linnaeus grouped all such fruits under the genus Citrus. It is believed that different groups of Citrus and related genera originated in multiple locations. For instance, Citrus sinensis is considered to have its origin in China, whereas Citron (Citrus medica) is known to have originated in Indonesia’s archipelago and North India [7][1]. Many Citrus fruits or their progenitors had their origin in northeast India [2], and Citrus indica is the most primitive and probable ancestor of the other Citrus species [8][3]. This observation was based on the discovery of Citrus indica in wild condition in forests of Nagaon district (Assam), Meghalaya, and Manipur in India. Citrus jambhiri is known to occur in wild conditions in the Siang river basin of Arunachal Pradesh [9][4].
Citruses and their relatives have their point of the origin in different regions of Asia and Australia [10][5]. The contribution of Swingle in the origin and diversification of Citrus is considered the main clue for the recognition of new Citrus varieties. Six species of Citrus were recognized; two were native to Papua New Guinea and four were native to Australia [3][6]. Eremocitrus genus (E. glauca) is native to the New South Wales and Queensland deserts (Australia). The Clymenia genus (C. polyandra) originated in the Papua New Guinea. Poncirus genus, with deciduous leaves, has its origin in northern China and is tolerant to freezing temperatures (resisting up to −20 °C). Papeda group includes species from different origins; Citrus micrantha (Southern islands of the Philippines), C. latipes (Northeast India), C. macroptera (New Caledonia), C. celebica (Indonesian islands), and C. hystrix (could be from the Philippines) [3][6].
A new species, namely, Poncirus (P. polyandra), was recognized, showing the properties of tolerating low temperatures, immunity against Citrus tristeza, and resistance to Phytophthora spp. [11][7]. It is directly used or crossed with other Citrus species to produce rootstocks for Citrus cultivation. The Citrus species is assumed to be derived from a large area in southeast Asia, probably mountainous regions of southern China and northeast India [12][8]; it was believed that Yunnan Province could be the origin due to its vast diversity of citruses, and the primary center of origin for the Citrus [13][9]. Several Citrus species, such as C. medica, C. limon, C. aurantifolia, C. maxima, C. aurantium, and C. sinensis, originated in the south due to a theoretical dividing line running from the northwestern border of India, above Burma, to the Yunnan province of China and then to the south of the island of Hainan [14][10]. Moreover, C. reticulata and other citruses originated north of it. Northeast (NE) India and the Malay and East Indian Archipelago are the origin of Citrons and pummelos [15][11]. The different types of citruses and their f origins are summarized in Table 1. The broad view of the world-wide origin of Citrus is presented in Figure 1. This covers the various species of Citrus that originated in different locations of the globe, from which they were then migrated and domesticated.
Table 1.
Origin of
| Species | Common Name | Center of Origin |
|---|---|---|
| Citrus reticulata Blanco. | Mandarin Orange | Philippines or Cochin China/Secondary center Japan |
| Citrus sinensis Pers. | Sweet Orange | China/Cochin China |
| Citrus macroptera Montr. | Satkara | Northeast Region of India |
| Citrus jambhiri Lush. | Rough Lemon | Possibly India |
| Citrus indica Tanaka | Indian Wild Orange (Memang Narang) | Northeast Region of India |
| Citrus limetta Lush. | Sarbati Lime | Tropical Asia |
| Citrus limon (L.) Burm. | Assam Lemon | East of Himalaya, North Myanmar, South China |
| Citrus medico L. | Citron | Indonesian Archipelago/North India |
| Citrus aurantifolia (Christm.) Swingle | Kagzi Lime | Indonesian Archipelago/North India |
| Citrus karna Raffin. | Karna Khatta | Eastern Region of India |
| Citrus aurantium L. | Sour Orange | Asia/Cochin China |
| Citrus megaloxycarpa Lush. | Sishuphal (Bartenga) | Northeast Region of India |
| Citrus grandis (L.) Osbeck | Pummelo | South East Asia |
| Citrus ichangensis Swingle | Ichang | Southwest, Central, Western China |
| Citrus assamensis Dutta and Bhatacharya | Ada Jamir | Northeast Region of India |
| Citrus latipes (Swingle) Tanaka | Khasi Papeda | Hills of Meghalaya and Nagaland |
| Citrus limonia Osbeck | Rangpur Lime | India and Sri Lanka |
The domestication of plants is the inherited custom of human nature. More than 1500 crops have been domesticated in the past [16][12]. The domestication of Citrus began independently in different parts of the world. The domestication of Citrus proceeded via the same trend as other plants, i.e., it took place in wild conditions before the domestication by humans began [17][13]. Interestingly, the currently available commercial varieties are mostly admixtures, which include oranges or lemons, cultivated back around two thousand years by the Romans. Evidence proves that the early cultivation of citron started in India, and of mandarins and other Citrus species, possibly in China. In Yunnan, wide Citrus diversity was observed [15][11] and in modern citriculture, crossbreeding varieties can partially recover the ancestral phenotypes of the introgressed genomes [18][14]. The migration of Citrus species via various other means is also a vital contributor to the diversity and domestication of different Citrus varieties. The development of early grafting techniques, such as revealed through the apomictic nature of some Citrus varieties, allowed for the rapid fixation of the genotypes [19][15]. This was because of the generation of seed clonal individuals, which explains the apomictic nature of commercial Citrus found at the current time [20][16].
2. Biodiversity
In general, the most marketed varieties worldwide are lemon, orange, mandarin, and grapefruit. Despite being the most crucial horticultural product due to their high level of hybridization during the domestication process, the recent taxonomic classification of Citrus remained unclear and unexplored. Because of its wide adaptability, Citrus rapidly spread and diversified in different parts of the world, though mainly in the West. As per the ancient records of Theophrastus, Citrus reached the Mediterranean region of Europe as early as 310 BC [21][17]. After dispersal, diversification occurred in the new localities. For instance, some genera of Aurantioideae viz. Afraegle, Aeglopsis, Balsamocitrus, and Citropsisare are native to tropical Africa, while genera such as Microcitrus, and Eremacitrus are native to Australia [22][18]. Hence, the overall origin of the Citrus genus is Southeast Asia, including South China, Northeast India, Burma. Most commercial cultivation of Citrus is undertaken through vegetative means to maintain quality and genetic uniformity. Moreover, Citrus is propagated through cutting and begins flowering and fruiting early. Citrus plants freely cross between genera, even among those that are closely related to one another. As a result, the plants are extremely heterozygous, and the hybrids created by inter- or intraspecific crosses show a significant amount of variation [23][19]. Another unique feature of Citrus is polyembryony, i.e., the formation of several embryos in the same seed. Pollination triggers embryo development from nucellar tissue apart from the normal zygotic embryo [24][20]. Nucellar embryos are more vigorous and have enormous significance in breeding and cultivation since such seedlings can be used as rootstock, which provides robust growth apart from retaining the original genetic complement. Some species of Citrus, such as Pummelo, Tangelo, and Clementine varieties of mandarin, are known to be mono-embryonic [15][11]. Other mono-embryonic Citrus types include Citrus indica, C. japonica, C. maxima, and C. medica. Because of polyembryony, monoembryony, and intercrossing, gene complement in Citrus is highly variable, and very often the distinction between varieties and species becomes blurred. Due to this, diversity assessment in Citrus is complex.
In one of the old records in Northeast (NE) India, there are 17 species and 52 varieties, and 6 probable hybrids [2]; in another report, 17 species and 52 varieties were found [25][21]. As per exhaustive account, there are 23 species and 68 varieties [26][22]; C. macroptera, C. juko, and C. serotina belong to the Papeda sub-genus. In NE India, many of the lesser-known citruses are grown and maintained as backyard crops in household gardens. This traditional practice contributes to the conservation of the species. Most Citrus is diploid with chromosome number 2n = 18. It has been found that the chromosome number of different Citrus species, and accordingly, species such as C. macroptera, C. limetta, C. karna, C. aurantium, C. ichengensis, and C. assamensis, are diploid [27][23]. However, some species have different ploidy levels, such as C. reticulata (18, 36; diploid and tetraploid, respectively), C. sinensis (18, 27, 36; diplod, triploid, and tetraploid, respectively), C. limon (18, 36), C. medica (18, 27), C. aurantifolia (18, 27), C. grandis (18, 36), and C. limonia (18, 27, 36). Citruses of the NE region comprise six groups and the citruses of NE India were classified as mandarin, orange, pummelo-grapefruit, acid, papeda, and other minor citruses [7][1]. The indigenous Citrus species of this region have distinctive and valuable genetic traits such as resistance to biotic and abiotic stress, distinctive aroma, flavor, etc. Hence, they can be considered genetic sources of useful traits for molecular and conventional breeding, as show in Table 2.
Table 2.
Useful traits of various
| Character | Adaptation | Species | Variety |
|---|---|---|---|
| Abiotic | Moisture stress and low fertility. | C. medica | Mithajora, soh-manong |
| Humid Tropics (very high rainfall) | C. assamensis | Ada jamir | |
| Drought resistance | C. jambhiri | Soh-myndong | |
| Wide adaptability for various types of soils and climates | C. medica f. limon | Patilebu | |
| Water logging and low lying condition | C. medica f. limon | Godha patilebu | |
| Stress conditions of soil and climate | C. aurantium | Karun-jamir | |
| High cold resistance | C. ichangensis, C. laptis |
Ketsa-shuphu | |
| Biotic | Resistance to greening, tolerant to psoriasis and exocortis virus | C. macroptera | Tith Kera |
| Resistance ton scab, canker, and gummosis | C. limon | Assam lemon | |
| Resistance to greening disease | C. indica, C. laptis | ||
| Special character | Flavor like ginger of Eucalyptus | C. assamensis | Ada jamir |
| Flavor like cardamom and more juicy | C. medica f. limon C. macroptera |
Patilebu, Panijamir, Joratenga, Elachi-lebu Satkara |
|
| Prolific bearing | C. medica f. limon C. reticulata |
Kata-jamir, Soh-synteng, Soh-kompriak, Soh-sanikar |
|
| Superior quality albedo (rind) | C. medica | Bira-jora | |
| Fruit quality superior for preparing pickles, chutney, and squash | C. medica f. aurantifolia, C. auruntium | Abhayapuri lime, Godha-huntera |
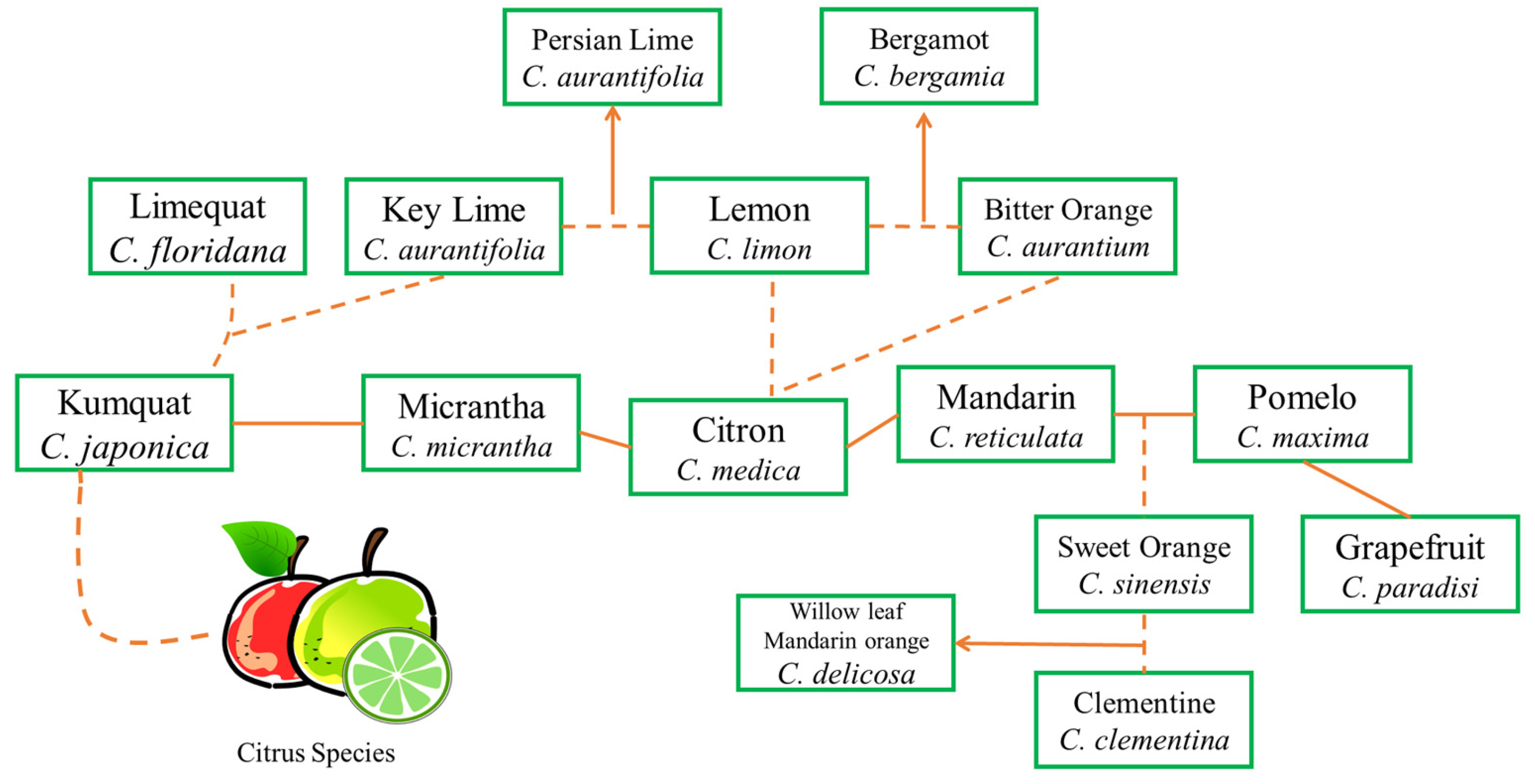
The domestication and diversification of Citrus reflects the status of Citrus and of the hybrids of various Citrus pure varieties (Figure 2). C. aurantium is a product of the natural hybridization of C. maxima and C. reticulata; in contrast, C. sinensis which is a second or third-generation product, is a direct hybrid or backcross between the ancestral taxa, i.e., (C. maxima × C. reticulata) × C. maxima [28][24]. C. clementina is a chance seedling hybrid (C. deliciosa × C. sinensis), discovered at end of the 19th century in Messerghin [27,29][23][25]. The genome is thus an admixture of C. reticulata and C. maxima; C. limon and C. aurantifolia are direct hybridization products of C. aurantium and C. medica and C. medica and C. micrantha, respectively [30][26]. New Caledonian and Kaghzi limes have an origin from F2 (C. micrantha × C. medica) × (C. micrantha × C. medica). Tahiti limes (seedless), C. latifolia (Bearss or IAC), are triploid hybrids of C. aurantifolia (diploid pollen) and C. limon (haploid ovule). Tanepao, Coppenrath, Ambilobe, and Mohtasseb limes, as well as triploid seedy limes and the Madagascar lemon, have evolved by hybridization of (C. micrantha × C. medica) × C. medica with a diploid gamete from the C. micrantha × C. medica.
Due to their independence from external influences, molecular markers such as SSR, SNP, and InDels [31][27] have greater advantages than phenotypic markers. InDel markers were discovered in a number of Citrus species [32][28]. It is difficult to assess how these genetic markers relate to physical and chemical characteristics [33][29]; hence, these markers might not explain variations in flavor profiles. In a recent study, a neighbor-joining method served to explain the diversity of Citrus. As ancestral species, citrus, mandarin, and pummelo represented a diversity pole; nonetheless, interspecific hybrids were found in close proximity [34][30]. Mosambi and Qicheng, Chanh Giay and Ma Nao Pan and Dalandan mandarins were shown to have close relationships; nevertheless, Pontianak and Nagpur were discovered to be intermediates to mandarins and pummelos. Based on SSR markers, Nagpur was previously described as a mandarin hybrid [35][31]. SSR [36][32], SRAP, and ISSR [37][33] were reported to exhibit low levels of polymorphism in oranges. Similarly, many hybrid accessions from mandarin × mandarin, mandarin × pummelo, or mandarin × tangelo have also been documented in mandarin data [38][34]. Citrons, rough lemons, and C. volkameriana were among the 56 accessions analyzed in citron using SRAP and SSR markers. The similarity levels were 0.70 (for citrons compared to other accessions) [39][35], 0.65 (for citrons and rough lemons) [40][36], 0.80 (for lemons and rough lemons compared to C. volkameriana) [39][35], and 0.79 (pummelo and grapefruit by ISSR) [41][37]. These studies highlight that genetic markers could be highly useful for the analysis of genetic variability among the Citrus varieties.
References
- Hore, D.K.; and Baruah, U. Status of citriculture in North Eastern Region of India—A Review. Agric. Rev. 2004, 25, 1–15.
- Barbhuiya, A.R.; Khan, M.L.; Dayanandan, S. Molecular phylogeny of Citrus species in the Eastern Himalayan region of Northeast India based on Chloroplast and Nuclear DNA sequence data. In Molecular Genetics and Genomics Tools in Biodiversity Conservation; Springer: Singapore, 2022; pp. 185–201.
- Devi, E.J.; Labala, R.K.; Sanabam, R.; Singh, N.S.; Modak, R.; Devi, H.S. New report of Citrus indica Yu. Tanaka, a wild progenitor species of citrus from Dailong Forest, Manipur, and recommendation for its conservation. Genet. Resour. Crop. Evol. 2022, 1, 545–558.
- Roy, A.; Das, S.K.; Tripathi, A.K.; Singh, N.U.; Barman, H.K. Biodiversity in North East India and their conservation. Progress Agric. 2015, 15, 182–189.
- Swingle, W.T. The Botany of Citrus and Its Relatives of the Orange Subfamily Aurantioidae of the Family Rutaceae; Webber, H.J., Batcheler, L., Eds.; University of California Press: Los Angeles, CA, USA, 1943.
- Swingle, W.T.; Reece, P.C. The botany of Citrus and its wild relatives. In The Citrus Industry; Reuther, W., Webber, H.J., Batchelor, L.D., Eds.; University of California Press: Berkeley, CA, USA, 1967; Volume 1, pp. 389–390.
- Zhang, S.; Chen, J.; Zhang, C.; Zhang, S.; Zhang, X.; Gao, L.; Yang, W. Insights into identifying resistance genes for cold and disease stresses through chromosome-level reference genome analyses of Poncirus polyandra. Genomics 2023, 115, 110617.
- Wu, G.A.; Terol, J.; Ibanez, V.; López-García, A.; Pérez-Román, E.; Borredá, C.; Domingo, C.; Tadeo, F.R.; Carbonell-Caballero, J.; Alonso, R.; et al. Genomics of the origin and evolution of Citrus. Nature 2018, 554, 311–316.
- Gmitter, F.G.; Hu, X. The possible role of Yunnan, China, in the origin of contemporary Citrus species (Rutaceae). Econ. Bot. 1990, 44, 267–277.
- Tanaka, T. Species Problem in Citrus (Revisioaurantiacearum, IX); Japan Society for the Promotion of Science: Tokyo, Japan, 1954; p. 152.
- Nicolosi, E.; Deng, Z.; Gentile, A.; Malfa, S.L.; Continella, G.; Tribulato, E. Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor. Appl. Genet. 2000, 100, 1155–1166.
- Meyer, R.S.; Purugganan, M.D. Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 2013, 14, 840–852.
- Wu, G.A.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Machado, M.A. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014, 32, 656–662.
- Terol, J. A whole genome association study in mandarin hybrids. In Proceedings of the Plant and Animal Genome XXVIII Conference, San Diego, CA, USA, 11–15 January 2020.
- Mudge, K.; Janick, J.; Scofield, S.; Goldschmidt, E.E. A history of grafting. In Horticultural Reviews; John Wiley & Sons: Hoboken, NJ, USA, 2009; Volume 35, pp. 437–439.
- Wang, X.; Xu, Y.; Zhang, S.; Cao, L.; Huang, Y.; Cheng, J.; Wu, G.; Tian, S.; Chen, C.; Liu, Y.; et al. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat. Genet. 2017, 49, 765–772.
- Kimball, D.A. Citrus Processing: A Complete Guide; Springer: Berlin/Heidelberg, Germany, 2012.
- Ulubelde, M. Turunçgillerintaksonomisi. Ege Bölgesi Zirai Arastırma Enstitüsü Yayınları 1985, 55, 43.
- Fatima, B.; Usman, M.; Khan, M.S.; Khan, I.A.; Khan, M.M. Identification of Citrus Polyploids using Chromosome Counts, Morphological and SSR Markers. Pak. J. Agric. Sci. 2015, 52, 107–114.
- Das, A.; Mondal, B.; Sarkar, J.; Chaudhuri, S. Genetic resource survey of mandarin orange (Citrus reticulata Blanco) in the northeastern Himalayan region of India. PGR Newsl. 2005, 143, 35–39.
- Chutia, M.; Bhuyan, P.D.; Pathak, M.G.; Sarma, T.C.; Boruah, P. Antifungal activity and chemical composition of Citrus reticulata Blanco essential oil against phytopathogens from North East India. LWT-Food Sci. Technol. 2009, 42, 777–780.
- Sharma, B.D.; Hore, D.K.; and Gupta, S.G. Genetic resources of Citrus of north—Eastern India and their potential use. Genet. Resour. Crop. Evol. 2005, 51, 411–418.
- Curk, F.; Ollitrault, F.; Garcia-Lor, A.; Luro, F.; Navarro, L.; Ollitrault, P. Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers. Ann. Bot. 2006, 117, 565–583.
- Federov. Chromosome Numbers of Flowering Plants; Otto Koeltz Science Publisher: Konigstein, Germany, 1974.
- García-Lor, A.; Luro, F.; Navarro, L.; Ollitrault, P. Comparative use of InDel and SSR markers in deciphering the interspecific structure of cultivated citrus genetic diversity: A perspective for genetic association studies. Mol. Genet. Genom. 2012, 287, 77–94.
- Ollitrault, P.; Curk, F.; Krueger, R. Citrus taxonomy. In The Genus Citrus; Woodhead Publishing: Sawston, UK, 2020; pp. 57–81.
- Le Nguyen, K.; Grondin, A.; Courtois, B.; Gantet, P. Next-generation sequencing accelerates crop gene discovery. Trends Plant. Sci. 2019, 24, 263–274.
- Noda, T.; Daiou, K.; Mihara, T.; Nagano, Y. Development of Indel markers for the selection of Satsuma mandarin (Citrus unshiu Marc.) hybrids that can be used for low-cost genotyping with agarose gels. Euphytica 2020, 216, 1–13.
- Yi, K.U.; Zhin, K.L.; Oh, E.U.; Kim, S.S.; Kim, H.B.; Song, K.J. Phenotypic and genetic characterization of three different types of Dangyooza (Citrus grandis), Korean landrace citrus. Hortic. Sci. Technol. 2021, 39, 96–105.
- Goh, R.M.; Pua, A.; Luro, F.; Ee, K.H.; Huang, Y.; Marchi, E.; Liu, S.Q.; Lassabliere, B.; Yu, B. Distinguishing citrus varieties based on genetic and compositional analyses. PLoS ONE 2022, 17, e0267007.
- Barkley, N.A.; Roose, M.L.; Krueger, R.R.; Federici, C.T. Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theor. Appl. Genet. 2006, 112, 1519–1531.
- Novelli, V.M.; Cristofani, M.; Souza, A.A.; Machado, M.A. Development and characterization of polymorphic microsatellite markers for the sweet orange (Citrus sinensis L. Osbeck). Genet. Mol. Biol. 2006, 29, 90–96.
- Fang, D.Q.; Roose, M.L. Identification of closely related citrus cultivars with inter-simple sequence repeat markers. Theor. Appl. Genet. 1997, 95, 408.
- Uzun, A.Y.; Yesiloglu, T.; Aka-Kacar, Y.I.; Tuzcu, O.; Gulsen, O. Genetic diversity and relationships within Citrus and related genera based on sequence related amplified polymorphism markers (SRAPs). Sci. Hortic. 2009, 121, 306–312.
- Uzun, A.; Yesiloglu, T.; Polat, I.; Aka-Kacar, Y.; Gulsen, O.; Yildirim, B.; Tuzcu, O.; Tepe, S.; Canan, I.; Anil, S. Evaluation of genetic diversity in lemons and some of their relatives based on SRAP and SSR markers. Plant. Mol. Biol. Report. 2011, 29, 693–701.
- Gulsen, O.; Roose, M.L. Lemons: Diversity and relationships with selected Citrus genotypes as measured with nuclear genome markers. J. Am. Soc. Hortic. Sci. 2001, 126, 309–317.
- Gulsen, O.; Uzun, A.; Canan, I.; Seday, U.; Canihos, E. A new citrus linkage map based on SRAP, SSR, ISSR, POGP, RGA and RAPD markers. Euphytica 2010, 173, 265–277.
More