Plasmid-mediated quinolone resistance (PMQR) remains one of the main mechanisms of bacterial quinolone resistance and plays an important role in the transmission of antibiotic resistance genes (ARGs). In this study, two novel plasmids, p3M-2A and p3M-2B, which mediate quinolone resistance in Proteus vulgaris strain 3M (P3M) were identified. Of these, only p3M-2B appeared to be a qnrD-carrying plasmid. Both p3M-2A and p3M-2B could be transferred into Escherichia coli, and the latter caused a twofold change in ciprofloxacin resistance, according to the measured minimum inhibitory concentration (MIC). Plasmid curing/complementation and qRT-PCR results showed that p3M-2A can directly regulate the expression of qnrD in p3M-2B under treatment with ciprofloxacin, in which process, ORF1 was found to play an important role. Sequence alignments and phylogenetic analysis revealed the evolutionary relationships of all reported qnrD-carrying plasmids and showed that ORF1–4 in p3M-2B is the most conserved backbone for the normal function of qnrD-carrying plasmids. The identified direct repeats (DR) suggested that, from an evolutionary
perspective, p3M-2B may have originated from the 2683-bp qnrD-carrying plasmid and may increase the possibility of plasmid recombination and then of qnrD transfer. To the best of our knowledge, this is the first identification of a novel qnrD-carrying plasmid isolated from a P. vulgaris strain of shrimp origin and a plasmid that plays a regulatory role in qnrD expression. This study also sheds new light on plasmid evolution and on the mechanism of horizontal transfer of ARGs encoded by plasmids.
- quinolone resistance
- Proteus vulgaris
- qnrD-carrying plasmids
- qnrD expression
- direct repeats
- homologous recombination
1. Effect of p3M-2A on the Expression of qnrD in p3M-2B
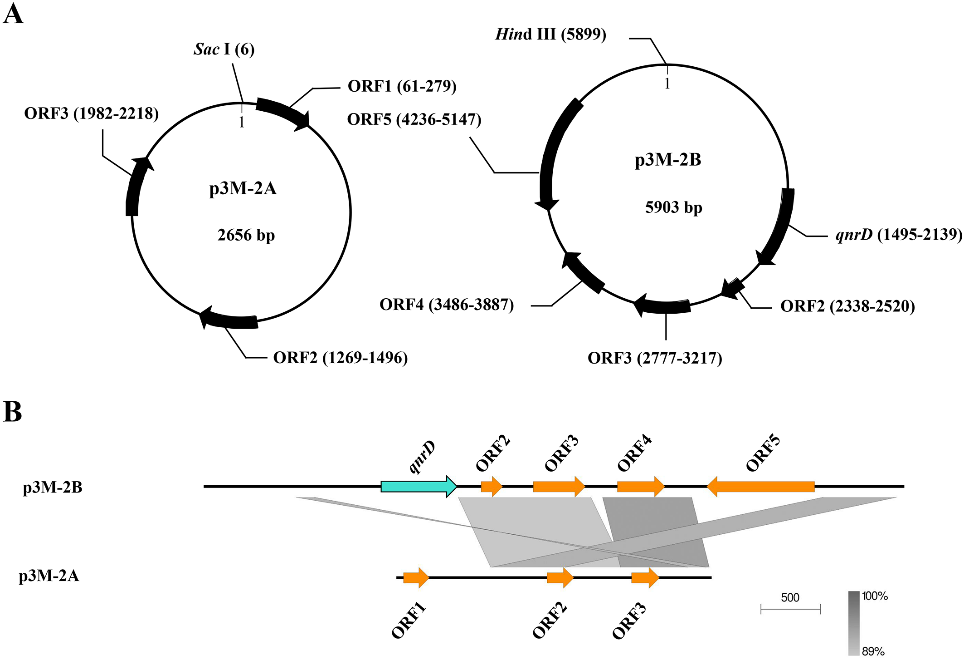
The plasmids p3M-2A and p3M-2B of P3M were sequenced, and their total length resulted to be 2656 bp and 5903 bp, respectively (Figure 1A). No resistance genes were present in p3M-2A, while p3M-2B carried qnrD. In this study, we found a certain correlation between these two plasmids—a large portion of the entire p3M-2A sequence shared a relatively high sequence identity (>89%) with ORF2–ORF4 and their intergenic regions of p3M-2B, implying that these two plasmids may originate from the same plasmid backbone (Figure 1B). Besides, we did not find any sequence with high identity withMicroorganisms ORF1 of p3M-2A in the database, and the same was observed for ORF5 of p3M-2B.
Figure 1. Graphical maps (A) and comparison of the structures (B) of p3M-2A and p3M-2B. The grey and dark shading in (b) indicates common regions between the plasmids.
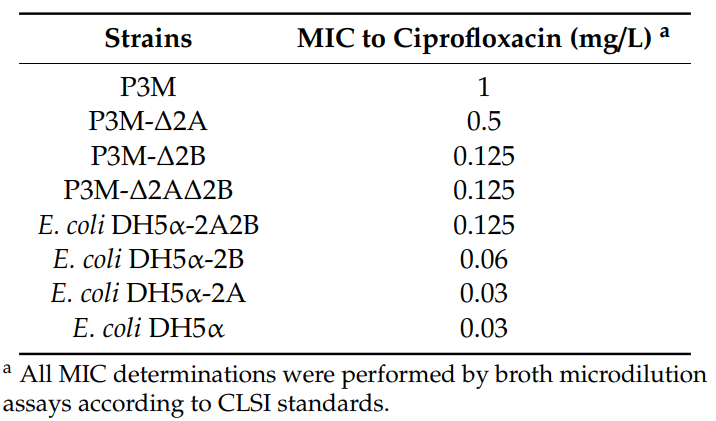
In order to verify the function of p3M-2A and p3M-2B, different plasmid deletion strains of P3M were generated: P3M-∆2A (p3M-2A elimination), P3M-∆2B (p3M-2B elimination), and P3M-∆2A2B (p3M-2A and p3M-2B elimination). In addition, p3M-2A and p3M-2B were transferred into E. coli DH5α separately or simultaneously to generate transformants with different plasmids: E. coli DH5α-2A (transformant containing p3M-2A), E. coli DH5α-2B (transformant containing p3M-2B), and E. coli DH5α-2A2B (transformant containing p3M-2A and p3M-2B). The ciprofloxacin MIC of these strains were determined. As shown in Table 2, ciprofloxacin resistance was decreased in P3M-∆2A and P3M-∆2B compared with P3M. Likewise, the resistance to ciprofloxacin was also reduced in E. coli DH5α transformants containing only p3M-2A or p3M-2B compared with E. coli DH5α-2A2B. These results showed that the presence of both plasmids caused an eight- or four-fold change in the ciprofloxacin resistance of P3M and E. coli DH5α-2A2B compared with the plasmid-free strains, respectively. In addition, we found that the ciprofloxacin resistance of P3M-∆2A and E. coli DH5α-2B strains containing only p3M-2B increased by four times or two times, respectively, while p3M-2A did not contribute to strain resistance. These results indicated that the two plasmids played synergistic roles in improving strain resistance to ciprofloxacin, and p3M-2B had a more obvious promoting effect. It is worth noting that, despite the absence of qnrD, p3M-2A still played a specific role in improving
the quinolone resistance of the bacteria tested.
Table 2. Determination of the MIC of ciprofloxacin in different transformants with respect to the parental strains.
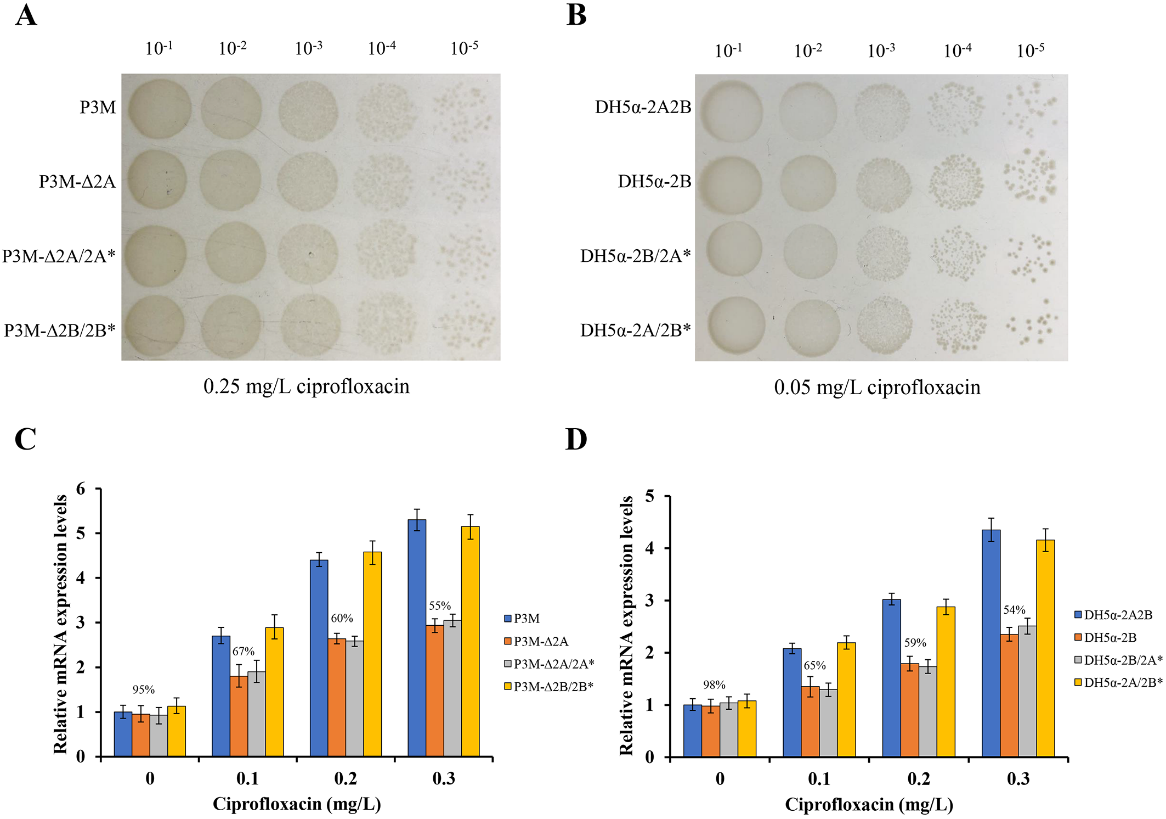
To confirm the function of p3M-2A, we carried out spot growth assays to test the effect of the absence of p3M-2A on the stability of p3M-2B. After subcultured for 100 generations, P3M-∆2A and DH5α-2B were able to preserve the ciprofloxacin-resistant phenotype in the presence of 0.25 mg/L and 0.05 mg/L of ciprofloxacin, respectively (Figure 2A,B), showing that p3M-2B could replicate and function independently and the absence of p3M-2A had no effect on the stability of p3M-2B in either P3M or E. coli DH5α strains. Considering that p3M-2A could improve the ciprofloxacin resistance of the strain even in the absence of qnrD, we speculated that this plasmid may have specific regulatory functions. Thus, we measured the expression level changes of qnrD in the presence or absence of p3M-2A using qRT-PCR (Figure 2C,D). The expression level of qnrD in P3M and P3M-D2A showed little difference in the absence of ciprofloxacin (Figure 2C). As the concentration of ciprofloxacin increased, the expression level of qnrD in both strains increased to different degrees; qnrD expression in P3M-D2A was apparently lower than that in the wild-type strain, and the difference became more obvious as the concentration increased. Similarly, the expression of qnrD showed the same changing trend in E. coli DH5α transformants containing p3M-2B and p3M-2A2B (Figure 2D). These results indicated that p3M-2A played a positive regulatory role in the expression of qnrD in p3M-2B, that is, the higher the ciprofloxacin concentration in the environment, the more obvious the regulatory effect of p3M-2A.
Figure 2. (A) Spot growth assays of wild-type (WT) P3M, p3M-2A deleted strain (P3M-∆2A), p3M-2A* complemented strain (P3M-∆2A/2A*), and p3M-2B* complemented strain (P3M-∆2B/2B*) on LB agar in the presence of 0.25 mg/L of ciprofloxacin. (B) Spot growth assays of DH5α transformant containing both plasmids (DH5α-2A2B), DH5α transformant containing p3M-2B (DH5α-2B), DH5α-2B strain complemented with p3M-2A* (DH5α-2B/2A*), and DH5α-2A strain complemented with p3M-2B* (DH5α-2A/2B*) on LB agar in the presence of 0.05 mg/L of ciprofloxacin. (C) Expression of qnrD in P3M, P3M-∆2A, P3M-∆2A/2A*, and P3M-∆2B/2B* with different ciprofloxacin concentrations. (D) Expression of qnrD in DH5α-2A2B, DH5α-2B, DH5α-2B/2A*, and DH5α-2A/2B* with different ciprofloxacin concentrations.
Since the sequence of ORF2–ORF3 in p3M-2A shares relatively high identity with that of p3M-2B, while ORF1 does not (Figure 1B), we speculated that ORF1 might play a certain regulatory role in qnrD expression. Hence, we constructed a new p3M-2A plasmid deleting ORF1 (p3M-2A*) and transferred it into P3M-∆2A and DH5α-2B to verify the effect of ORF1. As shown in Figure 2A and 2B, the new plasmid p3M-2A* did not affect the normal replication of plasmid p3M-2B in P3M-∆2A and DH5α-2B, and qnrD expression in the transformants was subsequently detected. As expected, p3M-2A* failed to restore the expression of qnrD to the wild-type level, but the expression was similar to that of the p3M-2A-deficient strain (Figure 2C,D). These results indicated that ORF1 in P3M-2A was key to regulate qnrD expression, as p3M-2A without ORF1 could not play its important regulatory function.
2. Phylogenetic Analysis of qnrD-Carrying Plasmids
By analyzing all the qnrD-carrying plasmids reported so far, we found that almost all of them were harbored by bacteria isolated from common environmental “reservoirs” like human and animal intestine, urinary tract, feces, water body, etc. 47 qnrD-carrying plasmids have been reported to date, of which most of them were isolated from Enterobacteriaceae, with the genus Proteus accounting for a relatively large proportion.
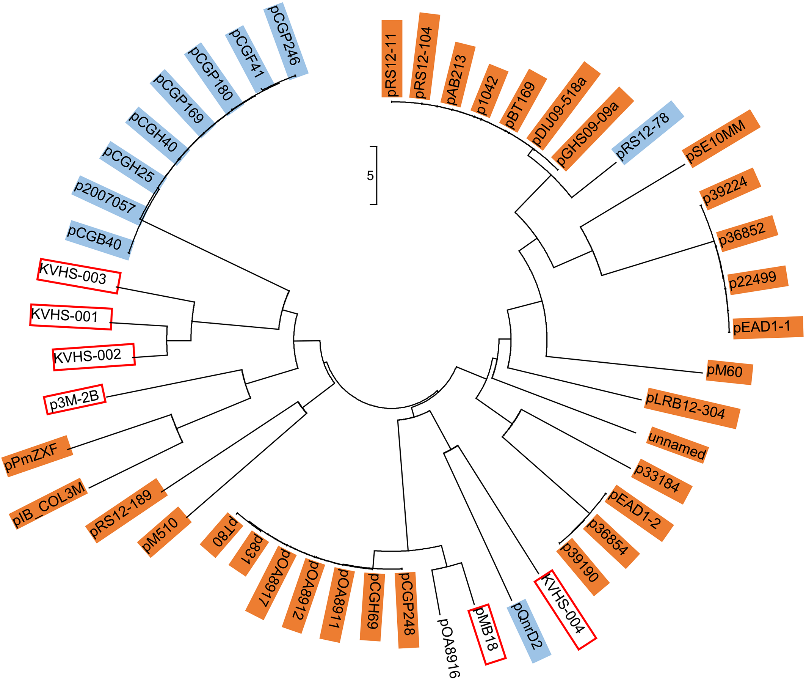
Almost all qnrD-carrying plasmids reported to date can be roughly divided into 2.7-kb and 4.3-kb categories according to their size. Interestingly, the p3M-2A plasmid in this study was about 2.7 kb in size, although it did not carry qnrD. The size of the p3M-2B carrying qnrD was 5.9 kb, and thus it did not comply with the above classification characteristics. In addition, we also found other six special cases that did not conform to the size rule: KVHS-001 (6.9 kb), KVHS-002 (8.1 kb), KVHS-003 (9.4 kb), KVHS-004 (6.2 kb), pOA8916 (2.0 kb), and pMB18 (5.2 kb) (Table S2). In order to further understand the genetic relationship between these qnrD-carrying plasmids, a neighbor-joining phylogenetic tree was constructed. As shown in Figure 3, plasmids of the same category tend to be more closely related to each other, while plasmids isolated from Proteus were dispersed and could be found in each clade, further illustrating the important function of Proteus species as disseminators in the phylogenetic evolution of qnrD-carrying plasmids.
Figure 3. Neighbor-joining phylogenetic tree of p3M-2B with other reported qnrD-carrying plasmids. Plasmids of 2.7 kb or 4.3 kb are highlighted in orange or blue, respectively. Plasmids larger than 4.3 kb are marked with red boxes. The evolutionary history was inferred using the neighbor-joining method. The optimal tree with the sum of branch length of 102.98104110 is shown. The tree is drawn to scale, with branch lengths measured in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and are expressed as the number of base substitutions per site. The analysis involved 49 nucleotide sequences. The codon positions included were 1st + 2nd + 3rd + Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 645 positions in the final dataset.
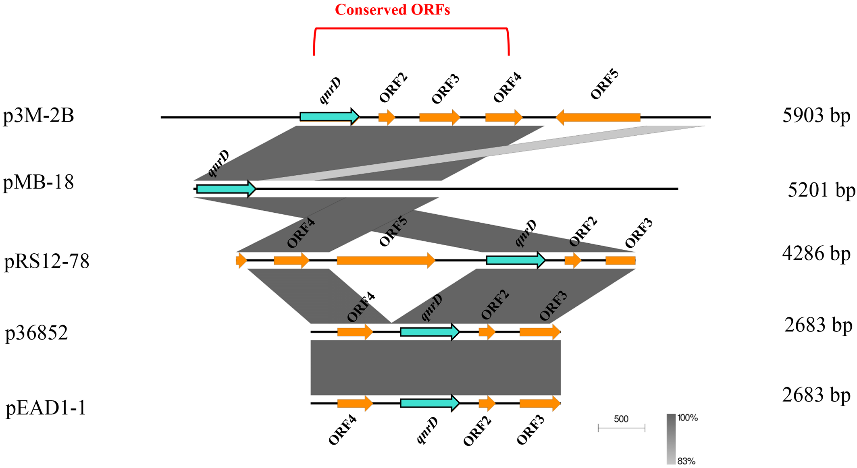
3. Possible Formation Process of p3M-2B
Due to the relatively large size of p3M-2B compared with the 2.7 and 4.3 kb qnrD-carrying plasmids, we hypothesized that it may have a specific formation process. We first focused on the other four plasmids isolated from P. vulgaris strains which were different in size and made sequence alignments. As shown in Figure 4, ORF1 (qnrD)-4 of p3M-2B was completely conserved in these four qnrD-carrying plasmids, with sequence identity up to 100%. Besides, the 2683-bp plasmids p36852 and pEAD1-1 appeared to be exactly composed of ORF1–4, implying that these four conserved genes of p3M-2B are the most conserved genes of the qnrD-carrying plasmids, necessary for their normal function.
Figure 4. Linear comparison of p3M-2B with other closely related qnrD-carrying plasmids isolated from P. vulgaris strains.
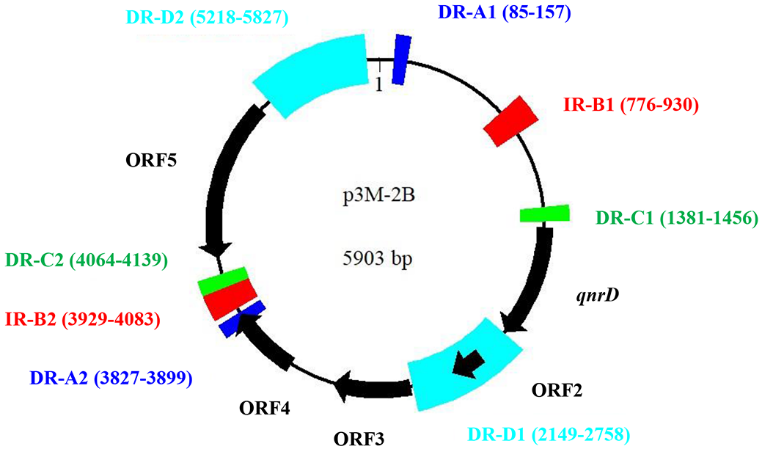
Subsequently, the p3M-2B plasmid sequence was analyzed with RepeatAround, a Windowsbased software tool designed to find repeats from 3 bp to 64 bp of length in circular genomes. We found that there were four pairs of DR or inverted repeats (IR) in p3M-2B (Figure 5), and it was notable that the sequence length between DR-C1 and -C2 was precisely 2683 bp, which fitted into the category of 2683-bp qnrD-carrying plasmids. In particular, the sequence between C1 and C2 corresponded exactly to ORF1–4, which appeared to be the most conserved and essential genetic component of qnrD-carrying plasmids for their proper function (Figure 4). It has been reported that DNA recombination mediated by DR regions is a major cause of genome plasticity, so it is conceivable that the DR present in p3M-2B would have a similar function. We hereby supposed that the 5903-bp p3M-2B may be originally derived from a 2683-bp plasmid, and DR-C1 and C2 play an important role in this process.
Figure 5. Graphical map of p3M-2B including direct repeats (DR) and inverted repeats (IR). Motifs of the same color represent a pair of DR or IR regions.
To test the above hypothesis, a new 2683-bp p3M-2B plasmid without ORF5 (p3M-2B*) was constructed, and its stability and qnrD expression in the transformants P3M-∆2B/2B* and DH5α-2A/2B* (Figure 2) were tested. Just as we expected, p3M-2B* was stable in the transformed strains (Figure 2A,B), and the expression of qnrD at different ciprofloxacin concentrations was not affected by ORF5 deficiency (Figure 2C,D). These results further indicated that ORF1–4 in qnrD-carrying plasmids is a very conserved backbone, and ORF5 in p3M-2B, which appears to be an acquired exogenous sequence, does not affect the normal replication process and further quinolone resistance.
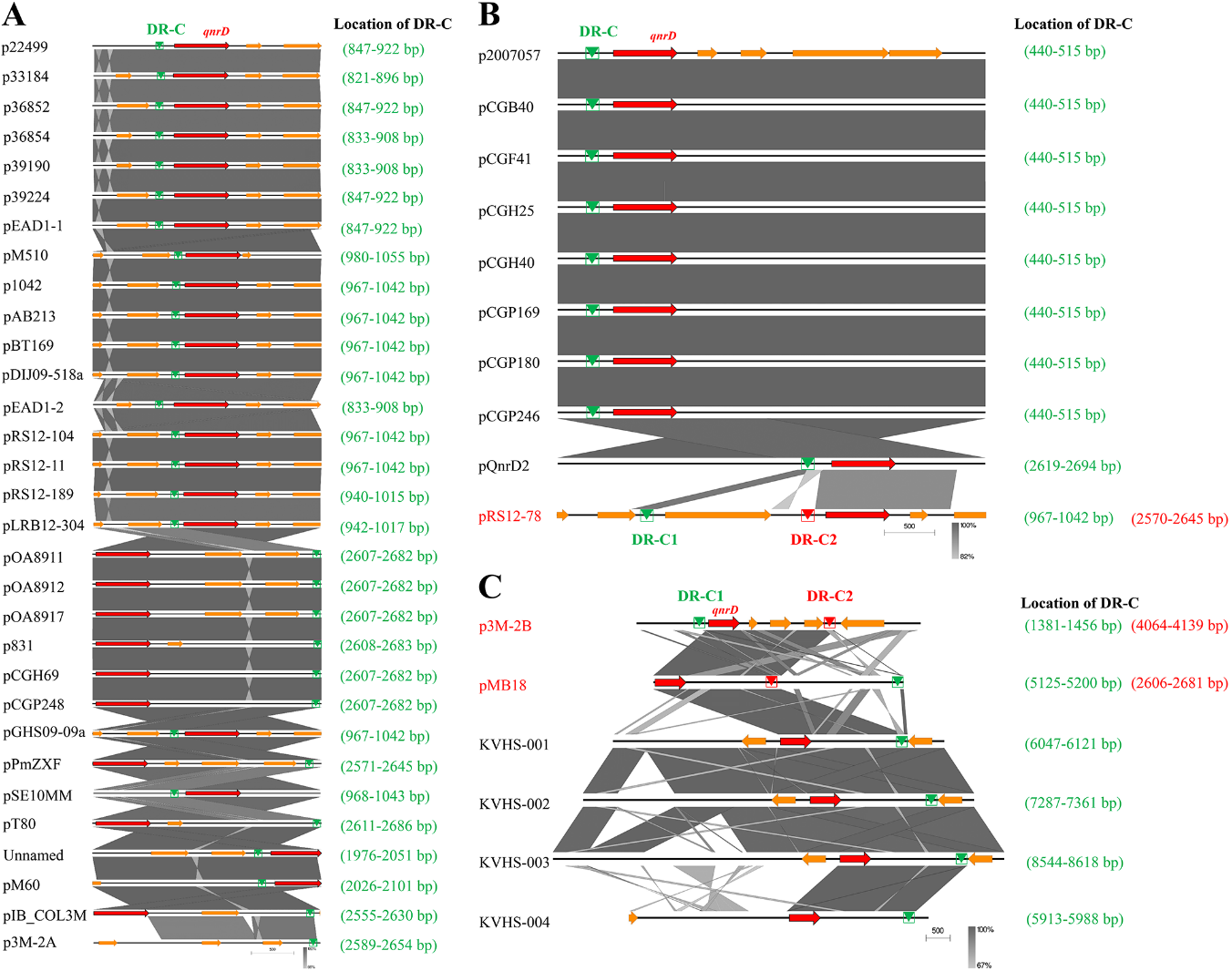
To further explore whether this is applicable to all larger qnrD-carrying plasmids, we made sequence alignments of the DR-C region in p3M-2B with similar regions in other reported qnrD-carrying plasmids. As shown in Figure 6, we found that although all plasmids contained DR regions sharing high sequence identity with DR-C, sequences varied slightly among different plasmids. The sequence with high identity to DR-C was present only in one site in the 2.7-kb plasmids, located upstream of qnrD (Figure 6A). It should be noted that although p3M-2A does not carry qnrD, it was also included in Figure 6A because it appeared to be in the 2.7-kb plasmid category and share high sequence identity with DR-C. Likewise, all 4.3-kb plasmids contained only one site identical with DR-C, with the exception of plasmid pRS12-78, containing two DR-C regions at different positions (Figure 6B). As shown in Figure 6C, among all plasmids larger than 4.3 kb, only pMB18 appeared to be consistent with p3M-2B, with two different DR regions (DR-C1 and C2), while only one DR-C was found in the four larger qnrD-carrying fragments isolated from Salmonella, which was unexpected. Plasmid pOA8916, smaller than 2.7 kb, contained one spot of DR-C upstream of qnrD as well (not shown).
In summary, among all the qnrD-carrying plasmids analyzed, only p3M-2B, pMB18, and pRS12-78 presented two different DR regions, and the sequence lengths between them resulted to be exactly 2683 bp, containing ORF1–4. Notably, these three plasmids were isolated from P. vulgaris strains, which hints at the important host role of P. vulgaris in plasmid propagation.
Figure 6. Comparative linear maps of qnrD-carrying plasmids of (A) 2.7 kb, (B) 4.3 kb, and (C) larger than 4.3 kb; qnrD and other predicted coding sequences (CDS) are denoted by red and orange arrows, respectively. The size of the arrows is to scale. The green and red triangles represent the regions sharing high sequence identity with DR-C in p3M-2B.
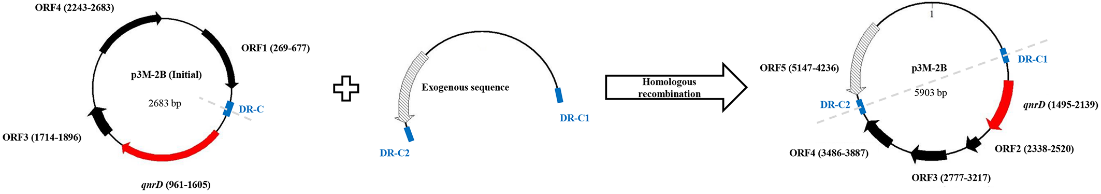
Based on the structural characteristics of the above three plasmids, we propose a potential model for the development of larger qnrD-carrying plasmids like p3M-2B (Figure 7). The p3M-2B backbone with an initial size of 2.7 kb contains DR-C region about 35–115 bp upstream of qnrD. The exogenous sequence containing DR-C at both ends in the adjacent environment may undergo homologous recombination with the DR-C region in p3M-2B (2.7 kb) and thus be integrated to turn into a new ORF. Ultimately, a novel plasmid p3M-2B of 5.9 kb is formed.
Figure 7. Possible model for in vivo formation of 5903-bp p3M-2B.
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms8071074