Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Pathology
Various bi-directional associations exist between oral health and gastro-intestinal diseases. The oral microbiome plays a role in the gastro-intestinal carcinogenesis and fusobacteria are the most investigated bacteria involved. The salivary microbiota can affect the development of the intestinal microbiota, as saliva flows through the gastrointestinal tract, allowing the bacteria present in it to easily reach the intestine. The DNA of bacteria belonging to the salivary microbiota is detectable in the gut even in low concentrations.
- oral microbiota
- Oral Health
- Gastro-intestinal Diseases
1. Oral Microbiota: An Overview
The mouth is the opening tract of the digestive system, and its unhealthy state has been bi-directionally associated with various systemic and gastro-intestinal diseases [1,2,3,4,5]. For example, atrophic glossitis and angular cheilitis may underlie Plummer–Vinson syndrome—a sideropenic dysphagia secondary to iron deficiency associated with gastric ulcerations [6] as well as pernicious anemia, which is due to the failure of the gastric cells to produce the intrinsic factor responsible for the absorption of vitamin B12 in the intestine [7]. Additionally, the oral manifestations of a number of chronic bowel diseases, such as Crohn’s and coeliac diseases, are well documented and oral signs and/or symptoms can be assumed to be related to these diseases [8,9,10].
The existence of an oral–gut axis has also been confirmed by the discovery of an association between intestinal bowel diseases (IBD) and pathogens of oral origin [11,12,13,14] as well as recent evidence that the administration of gut-derived probiotics can be useful in the prevention of dental caries [15]. This suggests that improving the condition of the gut microbiota may lead to a simultaneous improvement in the operational taxonomic units (OTUs) of bacteria residing in the oral cavity.
As defined by Berg et al. [16], the human microbiota is made up of all types of microorganisms (archaea, eukaryotes, bacteria and viruses), that live on and in the human body, each housed in specific ecological niches, including oral ones [17]. Instead, the term “microbiome” refers to all of their genomic material and products [18]. Ever since research into the oral microbiome began [19], new associations have been continuously identified between alterations in the composition of the oral microbiome and various gastrointestinal diseases. This is possible due to the development and use of high-throughput culture-independent technologies, such as reverse transcriptase-polymerase chain reaction (RT-PCR) [20] and next generation sequencing (NSG) [21], both capable of identifying microorganisms and their genes, even when they are not cultivable [22]. The “microbiome project” aims to identify the microbiome components of the entire human body [19] and the effects of various dysbiosis on human health. Approximately 700 species of prokaryotes have been identified in the oral microbiome. These species belong to 185 genera and 12 phyla, of which approximately 54% are officially named, 14% are unnamed (but cultivated) and 32% are known only as uncultivated phylotypes [9]. The 12 phlya are Firmicutes, Fusobacteria, Proteobacteria, Actinobacteria, Bacteroidetes, Chlamydiae, Chloroflexi, Spirochaetes, SR1, Synergistetes, Saccharibacteria (TM7) and Gracilibacteria (GN02) [23]. In addition, the oral cavity also contains diverse forms of microbes such as protozoa, fungi and viruses. Entamoeba gingivalis and Trichomonas tenax are the most commonly found protozoa and are mainly saprophytic. The Candida species is the most prevalent fungi seen associated with the oral cavity. Ghannoum et al. [24] carried out culture-independent studies on 20 healthy hosts and reported 85 fungal genera. The main species observed were those belonging to Candida, Cladosporium, Aureobasidium, Saccharomycetales, Aspergillus, Fusarium and Cryptococcus [25]. Of particular interest, Candida albicans is a dimorphic yeast that is occasionally found in healthy mouths as a saprophyte [26] but can infect oral mucosa after dysregulations of the normal oral flora, under local [27] and systemic circumstances, both para-physiological (pregnancies, elderly, early childhood) and iatrogenic (prolonged steroids and/or antibiotics therapies) [28] as well as in dysmetabolic/dysimmune pathologies (diabetes, obesity and/or immunodeficiencies) [29,30,31]. In all these situations, the antagonistic bacterial–fungal relationship favors the switch of C. albicans to its infectious phenotype, with carcinogenic potential, as has been reported for other microbial species such as papillomaviruses [32] in particular conditions [33].
Under physiological conditions, the microorganisms of the core microbiota are qualitatively and quantitatively arranged in three different ecological niches/intraoral habitats as follows: Group 1 includes microorganisms at the level of the keratinized gingiva, the hard palate and the buccal mucosa; Group 2 comprises those on the tongue, tonsils, throat (posterior wall of the oropharynx) and in the saliva; and Group 3 comprises those in sub- and supra-gingival plaque [34,35] (Figure 1).
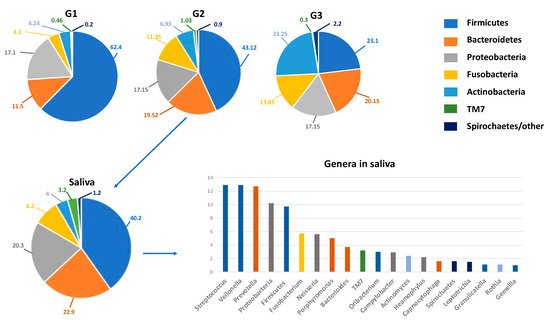
Figure 1. The oral microbiota of a healthy mouth. On top, the percentage composition of phyla in the various niches (G1–G3) of the oral microbiota. G1, Group 1: keratinized gingiva, hard palate and the buccal mucosa; G2, Group 2, tongue, tonsils, throat and saliva; G3, Group 3, sub- and supra-gingival plaque. On the bottom, details on the percentage composition of phyla (left) and genera mainly present in salivary microbiota (right). The genera are represented in descending percentages, and color bars reflect the phyla they belong to. The organisms inhabiting saliva account for 99.9% of all bacteria in the oral cavity and are usually planktonic organisms.
Oral dysbiosis consists of a qualitative and quantitative imbalance of the composition of the oral microbiota. The specific predominance of some pathogenic microorganisms over others is associated with specific oral and systemic diseases. Fusobacterium nucleatum and Porphyromonas gingivalis are the bacteria most frequently and variably investigated and are mainly known to be responsible for periodontal diseases, although they can also be found in subjects without periodontitis [36,37]. Scientific literature reports that their presence is associated with the onset and/or worsening of a wide range of systemic diseases [38], such as osteoporosis [5], cardiovascular diseases [39,40], rheumatoid arthritis [41] and neurodegenerative diseases such as Alzheimer’s and Parkinson’s [42,43].
2. Oral Dysbiosis and Gastrointestinal Diseases
The term “gastritis” refers to a series of acute or chronic inflammations of the stomach, secondary to endogenous or exogenous irritants, which determine a reparative and/or reactive response of the gastric mucosa. The etiology of acute gastritis (AC) recognizes numerous causes, such as drugs, caustic agents, radiations and traumas, while chronic gastritis (CG) is mainly sustained by Helicobacter pylori infections or autoimmunological triggers, and is dichotomized in H. pylori-related and H. pylori-unrelated gastritis [44]. The severity and persistence of inflammation in CG or the onset of ulcers, mainly related to H. pylori infections and non-steroidal anti-inflammatory drugs (NSAIDs), are associated with the risk of developing dysplasia of varying degrees and gastric cancers [45].
Furthermore, Cui et al. [46] analyzed the diversity of the oral microbiome of subjects with and without gastritis and found that the levels of 11 species decreased and 10 increased in patients with gastritis.
With regards to the gastro-intestinal system, F. nucleatum and P. gingivalis can reach the stomach directly during swallowing, but they and their toxins have also been found in the colon and in the fecal microbiome, because they pass gastric juices alive and by systemic dissemination from the ulcerated gingival pockets through the hematogenous route [47,48].
For these reasons, recent literature has focused on the hypothesis that gastric and colorectal cancers (CRC) and their precursors, such as gastritis and inflammatory bowel diseases (IBDs), may also be associated with oral dysbiosis [49,50]; furthermore, oral bacteria can colonize the gut microbiome, thus influencing intestinal and extra-intestinal health by altering the permeability of the intestinal mucosa and perpetuating chronic inflammatory states, both locally and systemically, with the hematogenous dissemination of lipopolysaccharides (LPSs) and other toxins responsible for distant consequences [51] as well as CRC and pancreatic cancers [49] (Figure 2).
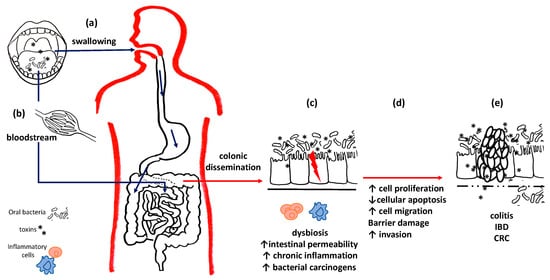
Figure 2. Oral bacteria and intestinal diseases. Oral bacteria can reach the intestines and stomach both through swallowing (a) and through the bloodstream (b). (c) At the level of the colon mucosa, they can compete with the local flora and establish intestinal dysbiosis. Pathogenic bacteria and their toxins alter the permeability of the basement membrane, perpetuate chronic inflammation and promote carcinogenesis. (d) Uncontrolled cell cycles reduced apoptotic efficiency and barrier damage results, thus leading to (e) chronic inflammatory diseases and the onset of cancers.
Zhang et al. [52] suggested three mechanisms of action of oral microbiota in the pathogenesis of cancer. The first is related to the state of chronic inflammation induced, especially by anaerobic species in particular, such as Porphyromonas, Prevotella and Fusobacterium. These pathogens are, in fact, able to stimulate the production of important mediators of the inflammatory process, such as the cytokines interleukin (IL)-1β, IL-6, IL-17, IL-23, and Tumor Necrosis Factor α (TNF-α). The consequent harmful effects develop mainly on fibroblasts, epithelial and endothelial cells, and components of the extracellular matrix, with an increase in the expression of the metalloproteases MMP-8 and MMP-9 and a consequent increase in cell proliferation, mutagenesis, initiation of angiogenic processes and oncogenesis [53,54].
A second mechanism of action involves the ability of oral bacteria to influence cell proliferation, rearrangement of the cytoskeleton, activation of the transcription factor NF-kB and inhibition of apoptosis. For example, it has been shown that P. gingivalis can inhibit apoptosis by influencing different pathways, reducing the expression levels of proapoptotic molecules such as p53 [54,55] and Bad [54,56], inhibiting the activation of caspase-9 [55] and increasing the production of anti-apoptotic factors, such as Bcl-2 [54,57].
The third mechanism involves the oral pathogens’ production of many carcinogenic substances. These include reactive oxygen (ROS) and nitrogen (RNS) species [58,59], mainly produced by species such as Streptococcus oralis, S. mitis, S. sanguinis, S. gordonii, S. oligofermentans [60], Lactobacillus fermentum, L. jensenii, L. acidophilus, L. minutus and Bifidobacterium adolescentis [61]. ROS and RNS production induces NADPH oxidase and nitric oxide synthase (NOS), respectively, along with their reactive oxygen and nitrogen species, which have been identified in various tumor types [62,63].
Other species, such as Porphyromonas gingivalis, Prevotella intermedia, Aggregatibacter actinomycetemcomitans and Fusobacterium nucleatum, produce volatile sulphur compounds (VSC), including hydrogen sulphide (H2S), methyl mercaptan (CH3SH), dimethyl sulphide ((CH3) 2S), (CH3SS), (CH3SS), and (CH3SS), whose presence is often associated with the onset of cancer [64,65,66].
Some species are able to produce more acids (e.g., Peptostreptococcus stomatis aciduric produces acetic, butyric, isobutyric, isovaleric, and isocaproic acids) [67] that can add to the acidic and hypoxic microenvironment of tumors, thereby increasing metastatic efficiency [68,69]. Furthermore, various oral microbial species, such as S. gordonii, S. mitis, S. oralis, S. salivarius and S. sanguinis streptococci [70], metabolize alcohol to acetaldehyde, which is indisputably carcinogenic.
Cordero et al. [71] reported that the relationship between oral hygiene and intestinal inflammation, which are mutually involved through signaling pathways, are linked to tumor-promoting inflammation. In fact, during the inflammatory process, the massive presence of pathogens or the simple imbalance of the oral microbiota play an important role in the onset of CRC from a chronic inflamed bowel, such as in cases of IBD. The authors based this hypothesis on the evidence that part of the gut microbiota comes from the oral one. Thus, as stated by Flemer et al. [49], “the oral microbiota in colorectal cancer is distinctive and predictive”. After profiling the microbiota from oral swabs, colonic mucosae and stools in individuals with CRC, colorectal polyps and healthy controls, they concluded that (i) oral bacteria were more abundant in CRC than in healthy controls, (ii) that the oral microbiome of healthy controls was different from those with CRC, and (iii) that F. nucleatum was most abundant in CRCs.
Numerous studies have also reported a significant increase in F. nucleatum in the gastric microbiome of subjects with gastric cancers and gastritis.
In 2013, Salazar et al. [72] conducted a clinical study to measure the levels of periodontal pathogenic bacteria in dental plaque and salivary samples from subjects with gastric precancerous lesions via quantitative RT-PCR. They found a high but not statistically significant increase in P. gingivalis and therefore, they hypothesized that high levels of periodontal colonization by pathogens may be associated with an increased risk of precancerous gastric lesions. In 2017, Coker et al. [73] identified differences in microbial diversity and richness between gastric cancers and various types of gastritis, thus indicating the presence of microbial dysbiosis in gastric carcinogenesis. Specifically, Prevotella intermedia and F. nucleatum, together with Prevotella oris and Catonella morbi, were significantly enriched in the gastric cancer microbiome compared to precancerous stages and they formed an increasingly strong co-occurrence network with disease progression.
The recent works of Yamamura et al. [74] and Hsieh et al. [75] have reinforced these correlations. Yamamura et al. [74] detected a significant increase in the mount of F. nucleatum DNA in oesophageal and gastric cancers as well as CRCs. Hsieh et al. [75] profiled gastric bacterial species in patients with gastritis and gastric cancer and found that F. nucleatum, along with Clostridium colicans, was frequently abundant in gastric cancer patients, supporting a specific gastric cancer signature.
Regarding C. albicans, the scientific literature has so far paid little attention to the study of its relationship with systemic health, limited to the role of candidiasis in a few systemic diseases, and has not yet considered its possible gastro-intestinal implications [76,77]. The main findings on oral bacteria associated with gastrointestinal disease are reported in Table 1.
Table 1. Oral bacteria associated with gastrointestinal diseases.
| G-i Diseases | Oral Bacteria/Fungi | Main Findings | Ref. |
|---|---|---|---|
| Gastritis | Streptococci, Fusobacteria | significantly higher in gastritis vs. healthy controls | [46] |
| Veillonella parvula, Corynebacterium matruchotii, Kingella oralis, Atopobium rimae, Aggregatibacter aphrophilus, Streptococcus sanguinis, Acinetobacter lwoffii, Prevotella amnii, Prevotella bivia, Cardiobacterium hominis and Oribacterium sinus | decreased in gastritis patients vs. healthy control | [46] | |
| Streptococcus infantis, Treponema vincentii, Leptotrichia unclassified, Campylobacter rectus, Campylobacter showae, Capnocytophaga gingivalis, Leptotrichia buccalis, Campylobacter concisus, Selenomonas flueggei and Leptotrichia hofstadii | increased in gastritis patients vs. healthy control (mainly Campylobacter spp.) | [46] | |
| Gastric Precancerous lesions | Campylobacter concisus | positively associated with the precancerous cascade of gastritis | [46] |
| Porphyromonas gingivalis, Treponema denticola | increased in dental plaque of subjects with gastric precancerous lesions | [72] | |
| Actinobacillus actinomycetemcomitans, Treponema denticola | significantly associated with gastric precancerous lesions | [72] | |
| Tannerella forsythia | significantly inversely associated with gastric precancerous lesions | [72] | |
| Oesophageal and Gastric Cancers | Tannerella forsythia, Porphyromonas gingivalis | associated with higher risk of oesophageal cancers | [52] |
| Streptococcus anginosus | higher in oesophageal cancer tissues than in oral cancer tissues | [52] | |
| Fusobacterium nucleatum | higher in oesophageal cancer tissues than matched normal mucosa; significantly associated with tumor stage and cancer-specific survival | [52] | |
| Neisseria spp., Candida glabrata | potential role in alcohol-related carcinogenesis |
[52] | |
| Parvimonas micra, Peptostreptococcus stomatis, Prevotella intermedia, Fusobacterium nucleatum, Prevotella oris, Gemella and Catonella morbi, Streptococcus anginosus, Dialister pneumosintes, Slackia exigua |
significantly increased in gastric cancer compared with precancerous stages |
[73] | |
| Inflammatory bowel diseases (IBDs) | Bacteroidetes | significantly increased in IBDs | [51] |
| Proteobacteria and Actinobacteria | increased in IBDs | [51] | |
| Campylobacter concisus | increases the mucosal permeability by affecting the tight junctions in IBDs | [51] | |
| Fusobacterium nucleatum | overrepresented in IBDs | [51] | |
| Candida albicans | isolated from the intestine more frequently in IBD patients | [51] | |
| CRC | Haemophilus spp., Prevotella spp., Alloprevotella Lachnoanaerobaculum, Neisseria and Streptococcus spp. |
less abundant in CRC than healthy controls | [49] |
| Fusobacterium nucleatum, Parvimonas micra, Peptostreptococcus stomatis, Dialister pneumosintes | tumor-associated bacteria | [49] | |
| Peptostreptococcus, Parvimonas, Fusobacterium | more abundant in CRC than in healthy controls | [49] | |
| Fusobacterium nucleatum | induces inflammatory response and promotes CRC development |
[52] | |
| Treponema denticola, Prevotella intermedia | increases the CRC risk | [52] | |
| Porphyromonas gingivalis | causes inflammation and promotes tissue degenerative processes | [52] | |
| Fusobacterium nucleatum | associated with CRC regional lymph node metastases | [55] | |
| Fusobacterium nucleatum, Selenomonas, Prevotella, Parvimonas micra, Peptostreptococcus stomatis | increased in CRC; induces colon cancer growth and progression | [71] | |
| Lachnospiraceae | can protect against CRC | [71] | |
| Fusobacterium nucleatum | sustains both the biofilm and the CRC tumorigenesis |
[47] |
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms9051064
This entry is offline, you can click here to edit this entry!
