We previously demonstrated that transplacental gene delivery (TPGD) method combined with CRISPR/Cas9 system can manipulate fetal genome in situ. Recently, we examined whether the endogenous gene (myosin heavy-chain α,
MHCa
) can be genome-edited by TPGD in the absence of a gene delivery reagent with potential fetal toxicity. For this, we employed a hydrodynamics-based gene delivery (HGD) system with the aim of ensuring biosafety. We also investigated which embryonic stages are suitable for the induction of genome editing in fetuses. As the results, it was indicated that the HGD-based TPGD of a genome editing vector enables to manipulate the fetal genomes.
- CRISPR/Cas9
- fetuses
- genome editing
- hydrodynamics-based gene delivery (HGD) system
- myosin heavy-chain α
- transplacental gene delivery (TPGD)
- TPGD for acquiring genome-edited fetuses (TPGD-GEF
1. TPGD
TPGD
Transplacental gene delivery, termed "TPGD" by our group
Transplacental gene delivery, termed “TPGD” by our group
[1]
, was first developed by Tsukamoto et al. with gene delivery accelerating reagent
[2]
, since then, several researchers have reported this system works well
. We intravenously introduced an all-in-one plasmid using the lipid-based gene delivery reagent, enabling the potential expression of both Cas9 and guide RNA (gRNA) targeted to enhanced green fluorescent protein (
EGFP
) cDNA into pregnant mice at E12.5
[7]. Molecular analysis of the fetus hearts showing the presence of insertion/deletion (indel) mutations at the target locus, although normal cells were also present. These findings suggested that TPGD-based genome editing is functional in mid-gestational fetuses. This is the first report that the TPGD is useful for inducing genome editing in fetus. We thus termed this technology "TPGD for acquiring genome-edited fetuses (TPGD-GEF)".
. Molecular analysis of the fetus hearts showing the presence of insertion/deletion (indel) mutations at the target locus, although normal cells were also present. These findings suggested that TPGD-based genome editing is functional in mid-gestational fetuses. This is the first report that the TPGD is useful for inducing genome editing in fetus. We thus termed this technology “TPGD for acquiring genome-edited fetuses (TPGD-GEF)”.
HGD-based TPGD-GEF
2. HGD-based TPGD-GEF
In the case of TPGD, to ensure fetal gene delivery, many researchers have employed gene delivery reagents such as lipids, liposomes, and polyethylenimines (PEIs), which can all protect DNA from degradation, enable the DNA to pass through the BPB smoothly, and increase the transfection efficiency in fetal cells
In the case of TPGD, to ensure fetal gene delivery, many researchers have employed gene delivery reagents such as lipids, liposomes, and polyethylenimines (PEIs), which can all protect DNA from degradation, enable the DNA to pass through the BPB smoothly, and increase the transfection efficiency in fetal cells
[1]
. However, these gene delivery reagents are often toxic to fetuses and sometimes cause fetal malformation
[4][8][9][10]. Considering the safety of the fetus, there is a need to develop TPGD without the use of such reagents.
. Considering the safety of the fetus, there is a need to develop TPGD without the use of such reagents.
Results: Analysis of HGD-Based TPGD-GEF
3. Results: Analysis of HGD-Based TPGD-GEF
In this study, we examined two points: the first was whether it was possible to disrupt an endogenous gene by this technology, and the second was whether it was possible to perform TPGD-GEF in the absence of a gene delivery reagent. For the first, we targeted endogenous myosin heavy-chain α (
MHCa
) gene in the heart. And for the second, we employed a hydrodynamics-based gene delivery (HGD) system. HGD relies on the introduction of a large volume of DNA solution at once without any gene delivery reagents and is a powerful method for the efficient transfection of murine hepatocytes
[11]
. Based on the original study, we used this technology with naked DNA for avoid the potential cytotoxicity. As the results, indel occurrence was observed at the endogenous
MHCa
locus in some of the fetuses carrying pCGSap1-
MHCa,
which is an all-in-one plasmid conferring the expression of both Cas9 and guide RNA (targeted to the
MHCa
), when HGD-based TPGD-GEF was performed on pregnant females at E9.5. Of the three pregnant females injected, one had mutated fetuses: all examined fetuses carried exogenous plasmid DNA, and four of 10 (40%) exhibited mosaic indel mutations in
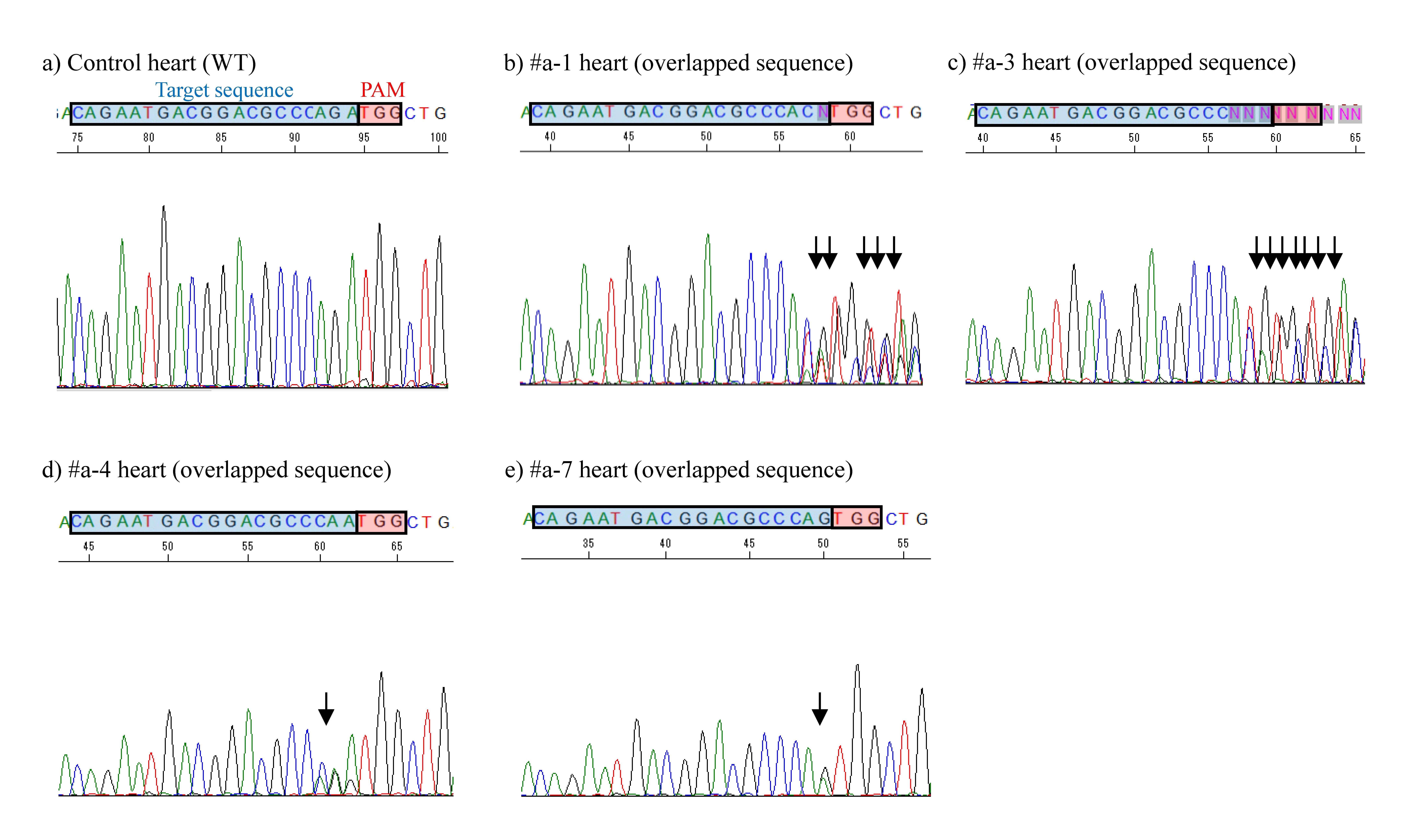
MHCa. Although the mode of mutation in the obtained fetuses was mosaic as shown by the presence of edited and unedited cells (Figure 1). Gene delivery to fetuses at E12.5 and E15.5 did not cause mutations.
. Although the mode of mutation in the obtained fetuses was mosaic as shown by the presence of edited and unedited cells (Figure 1). Gene delivery to fetuses at E12.5 and E15.5 did not cause mutations.
Figure 1
. Sequence analysis of PCR products (corresponding to the endogenous
MHCa
gene exon 1) derived from HGD-based TPGD-GEF-treated fetuses. Direct sequencing of PCR products obtained from fetal hearts (fetus of WT and fetuses of #a-1, -3, -4, and -7). In a) control heart, the sequence enclosed by a quadrant filled with blue (corresponding to the target sequence recognized by gRNA targeting MHC
a sequence) remained unaltered. By contrast, in b) #a-1, c) #a-3, d) #a-4, and e) #a-7, overlapping electrophoretograms (indicated by arrow) are notable immediately upstream of the PAM, indicating the presence of genome-edited and unedited sequences. The PAM (TGG) is shown as a quadrant filled with red. WT, wild-type.
sequence) remained unaltered. By contrast, in b) #a-1, c) #a-3, d) #a-4, and e) #a-7, overlapping electrophoretograms (indicated by arrow) are notable immediately upstream of the PAM, indicating the presence of genome-edited and unedited sequences. The PAM (TGG) is shown as a quadrant filled with red. WT, wild-type.
4. Conclusion
Conclusion
This study
This study
[12]
has novelty, which is clearly different from other similar research including our previous paper in view that: 1) successful genome editing was achieved when HGD was employed without using any gene delivery reagents, and 2) mutations are induced successfully at the target endogenous
MHCa
locus. In addition, against our expectations, cells with genome editing were found in cardiac cells, and in other non-cardiac cells. E9.5 appears to be the appropriate stage for manipulating the function of embryonic cells by the HGD-based TPGD-GEF, although there is still room for improvements to this system.
References
- Shingo Nakamura; Satoshi Watanabe; Naoko Ando; Masayuki Ishihara; Masahiro Sato; Transplacental Gene Delivery (TPGD) as a Noninvasive Tool for Fetal Gene Manipulation in Mice. International Journal of Molecular Sciences 2019, 20, 5926, 10.3390/ijms20235926.
- Makoto Tsukamoto; Takahiro Ochiya; Sho Yoshida; Takashi Sugimura; Masaaki Terada; Gene transfer and expression in progeny after intravenous DNA injection into pregnant mice. Nature Genetics 1995, 9, 243-248, 10.1038/ng0395-243.
- N Kikuchi; S Nakamura; M Ohtsuka; M Kimura; M Sato; Possible mechanism of gene transfer into early to mid-gestational mouse fetuses by tail vein injection. Gene Therapy 2002, 9, 1529-1541, 10.1038/sj.gt.3301818.
- Rui Maeda-Mamiya; Eisei Noiri; Dr. Hiroyuki Isobe; Waka Nakanishi; Koji Okamoto; Kent Doi; Takeshi Sugaya; Tetsuro Izumi; Tatsuya Homma; Eiichi Nakamura; et al. In vivo gene delivery by cationic tetraamino fullerene. Proceedings of the National Academy of Sciences 2010, 107, 5339-5344, 10.1073/pnas.0909223107.
- A. M. Efremov; A. O. Buglaeva; Sergey Orlov; Sergey Burov; I. A. Ignatovich; E. B. Dizhe; Vladimir S. Shavva; Andrej Perevozchikov; Transfer of genetic constructions through the transplacental barrier into mice embryos. Russian Journal of Developmental Biology 2010, 41, 71-76, 10.1134/s1062360410020025.
- Ning Wu; Ai-Bing Yu; Hua-Bin Zhu; Xiu-Kun Lin; Effective Silencing of Sry Gene with RNA Interference in Developing Mouse Embryos Resulted in Feminization of XY Gonad. Journal of Biomedicine and Biotechnology 2012, 2012, 1-11, 10.1155/2012/343891.
- Shingo Nakamura; Masayuki Ishihara; Naoko Ando; Satoshi Watanabe; Takayuki Sakurai; Masahiro Sato; Transplacental delivery of genome editing components causes mutations in embryonic cardiomyocytes of mid-gestational murine fetuses. IUBMB Life 2019, 71, 835-844, 10.1002/iub.2004.
- A V Bespamiatnova; S D Zaugol'nikov; Iu Z Sukhov; [Embryotoxic and teratogenic effect of ethyleneimine].. Farmakologiia i toksikologiia 1970, 33, 357-360.
- Mario C Filion; Nigel C Phillips; Toxicity and immunomodulatory activity of liposomal vectors formulated with cationic lipids toward immune effector cells. Biochimica et Biophysica Acta (BBA) - Biomembranes 1997, 1329, 345-356, 10.1016/s0005-2736(97)00126-0.
- Irving L.M.H. Aye; Jeffrey A. Keelan; Placental ABC transporters, cellular toxicity and stress in pregnancy. Chemico-Biological Interactions 2013, 203, 456-466, 10.1016/j.cbi.2013.03.007.
- F Liu; Y K Song; D Liu; Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Therapy 1999, 6, 1258-1266, 10.1038/sj.gt.3300947.
- Shingo Nakamura; Naoko Ando; Satoshi Watanabe; Eri Akasaka; Masayuki Ishihara; Masahiro Sato; Hydrodynamics-Based Transplacental Delivery as a Useful Noninvasive Tool for Manipulating Fetal Genome. Cells 2020, 9, 1744, 10.3390/cells9071744.