1. Introduction
Grapevine is one of the major fruit crops worldwide, in terms of economic value and cultivated area. A large amount is subjected to wine production; however, grape has become a model for molecular biology, genetics, and breeding, garnering significant research interest in different areas from the study of polyphenols
[1][2][3][1,2,3] to the investigation of specific gene families, such as resistant genes
[4].
Grapevine is a plant rich in bioactive compounds
[5][6][5,6], known for its therapeutic effects. The grape extract was studied for a long time for its hepatoprotective
[7], hypoglycemic
[8], cardio-protective
[9], antioxidant
[10][11][10,11], antibacterial
[11][12][13][11,12,13], and antiviral activity
[11][14][11,14] which are due to the high levels of polyphenolic compounds found in grapes skin, seeds, and stem. The anti-cancer effect, that has long been studied over the years, is attributable to the consistent antioxidant activity of these molecules
[15][16][15,16]. Resveratrol, for example, one of the most abundant molecules in the extracts, acts at various levels of carcinogenesis
[17]. Grape seed extract inhibits the expression of the epidermal growth factor receptor (EGFR) in head and neck cutaneous squamous cell carcinoma
[18], and of MEPK/ERK1-2 and MAPK/p38 in breast cancer, counteracting tumor invasiveness and progression. Some phenolic compounds, such as quercetin and catechins, given their similarity to estrogenic hormones, determine a pro- and anti-estrogenic receptor response in breast cancer
[19]. Sharma et al. demonstrated the apoptotic activity of proanthocyanidins blocking COX-2 and the prostaglandin receptor (PGE2)
[20]. In this context, two stilbenoids of wine, trans-astringin and trans-piceatannol, also show a cancer-chemiopreventive action by inhibiting cyclooxigenases and precancerous lesions
[21]. Recently, Di Meo et al. evaluated the anti-cancer activity of grape seeds semi-polar extracts of two Italian grape varieties (Aglianico and Falanghina). They found that the extracts of both varieties are able to inhibit growth and migration of three different mesothelioma cell lines by activating apoptosis
[22].
Antibacterial activity has widely been reported
[23][24][25][26][27][28][23,24,25,26,27,28]. Andelkovic et al.
[29] demonstrated that flavonols, phenolic acids, and flavan-3-ols were the main components of grape leaf extracts driving antioxidant and antibacterial activity, principally inhibiting Gram-positive bacteria. A similar effect has been observed by Rodriguez–Vaquero et al.
[30], who analyzed the antimicrobial potential of three Argentinian wines: Listeria monocytogenes growth was highly reduced by caffeic acid, rutin, and quercetin. Instead, Papadopoulou et al.
[31] reported that alcohol-free red and white wine extracts also had antimicrobial potential against Escherichia coli and Candida albicans, albeit it was less strong than that observed against Staphylococcus aureus.
2. Antiviral Activity of Vitis vinifera Leaf Extract against SARS-CoV-2 and HSV-1
2.1. Flavonoid Composition of V. vinifera Leaves
Negative ionization mode HPLC-MS/MS is one of the powerful methods to characterize flavonoids because it provides information on both aglycones and glycosidic conjugates that can differentiate isomers
[32][33][47,52]. HPLC-MS/MS IDA analysis was carried out to investigate V. vinifera leaves metabolic profile.
MZmine
[34][53] and GNPS
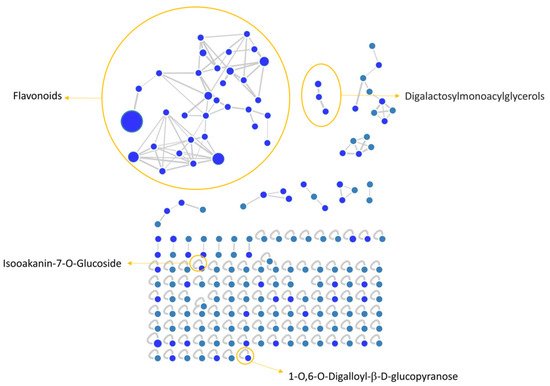
[35][54] software were used to convert and analyze the raw MS data, and finally Cytoscape built the complex network shown in
Figure 1. Each node indicates a precursor ion, the node size is strongly related to its abundance in the sample (sum precursor intensity), and the edges size between two nodes is directly dependent on their MS/MS spectra similarity (cosine score)
[36][55]. Analysis of GNPS results together with data manual curation evidenced the presence in the network of one big flavonoids cluster together with a smaller one belonging to digalactosylmonoacylglycerol (DGMG) lipids; other unpaired nodes have also been identified during GNPS analysis (
Figure 1).
Figure 1. V. vinifera extract Feature Based Molecular Network (FBMN) obtained through GNPS analysis and Citoscape visualization, displaying the presence of flavonoids and digalactosylmonoacylglycerol clusters, together with two unpaired nodes matching with 1-O,6-O-Digalloyl-ß-D-glucopyranose and Isookanin-7-O-glucoside.
Assignment of each node has been manually curated and the putative identification is reported, Results showed that compound 1 was not in a cluster and it was identified by the GNPS annotation tool as the tannin 1-O,6-O-Digalloyl-β-D-glucopyranose.
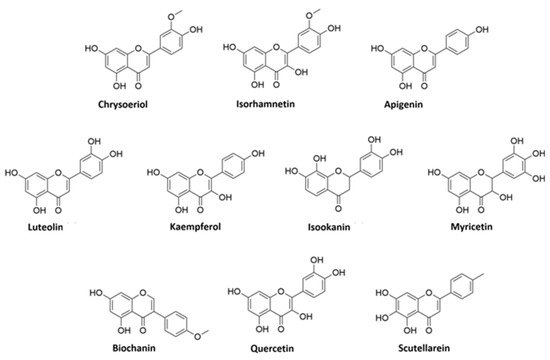
A detailed analysis of the MS and MS2 spectra, combined with the GNPS annotation tools, lead to the characterization of 35 flavonoids conjugates. Most of them were quercetin derivatives (8 compounds), others included derivatives of luteolin (5 compounds), kaempferol (4 compounds), apigenin (3 compounds), isorhamnetin (2 compounds), myricetin, chrysoeriol, biochanin, isookanin, and scutellarein (1 compound) (Figure 2). Four flavonoids were not identified (N.I.).
Figure 2. Structures of flavonoid aglycones identified in V. vinifera.
Moreover, we calculated the relative abundance of each compound present in Table 2 within the same chemical class. Results showed that compound 13 is the most representative among polyphenols, accounting for more than 44% of the total, followed by compounds 6 and 11, with 16% and 13%, respectively. Compounds 8, 14, 15, 26, and 28 account for less than 10% each, while all the others are present in less than 1% or traces . Among DGMGs, compound 37 is the most representative, with 97%, while compound 38 is present in traces and compound 39 represents 3% of the cluster.
2.2. Identification of Kaempferol and Luteolin Derivatives
The aglycone fragment at m/z 285 of compounds
2,
20,
23,
25, and
26 suggested that they originated from kaempferol or luteolin. Compounds
2,
20, and
26 produced a pseudo molecular ion at m/z 447 [M − H]
−, and the MS
2 profile showed typical fragments of hexose glycoside flavone, as they exhibited peaks at m/z 357 [M − H-90]
−, 327 [M − H-120]
−, and 297 [M − H-150]
−. Thus, based on the MS
2 fragmentation profile and RT, compound
2 could be assigned as kaempferol-8-C-glucoside, compound
20 as kaempferol-7-O-glucoside, and compound
26 as kaempferol-3-O-glucoside
[33][37][38][52,56,57].
Compound
23, m/z 593, is characterized by the neutral loss (NL) of (rhamnosyl-hexoside) moiety [M − H-308]
−, and by comparison of its MS
2 pattern with the literature, we assigned the identity as kaempferol-3-O-(6-O-rhamnosyl-galactoside)
[39][58]. Compound
25 showed the NL of a glucuronide unit [M − H-176]
− and it was assigned as kaempferol-3-O-glucuronide.
Compounds
4,
6,
8,
9, and
18 are characterized by the presence of aglycone fragment at m/z 285 and they are all isomers with a parental ion at m/z 447 [M − H]
−. These isomers were identified through the GNPS database and discriminated by RT as orientin, isoorientin, luteolin-4-O-glucoside, and ymaroside, respectively. Differently, compound
24 showed parental ion at m/z 483 and was generally assigned as luteolin-C-hexoside, by comparison with a precedent study
[40][59].
2.3. Identification of Apigenin Derivatives
Compounds
10,
11, and
29 exhibited the same [M − H]
− ion at m/z 431. Compounds
10 and
11 showed a typical fragment of C-glycosides [M − H-120]
− at m/z 311. In addition, their MS
2 pattern corresponds to the vitexin and isovitexin fragmentation profiles, respectively
[39][58]. The isomer
29 was generally assigned as apigenin-C-glucoside.
2.4. Identification of Quercetin Derivatives
Compounds
13,
14,
17,
19,
21,
22,
31, and
33 are all characterized by the presence of the quercetin aglycone at m/z 301. Compounds
13 and
17 showed parental ion at m/z 477 [M − H]
− with a NL of 176 Da [M − H-176]
− that is characteristic of the glucuronyl moiety. Compound
17 was identified by the GNPS assignment tool as quercetin-3-O-glucuronide, while its isomer, compound 13, was tentatively identified as quercetin-O-glucuronide
[41][60]. Compounds
14,
22, and
31 showed the same pseudomolecular ion at m/z 463 [M − H]
−, showing a NL of a glucoside moiety [M − H-162]
−. GNPS assigned compound 14 as isoquercitrin, while compounds
22 and
31 were tentatively assigned as hyperoside and quercetin-7-O-glucoside, respectively, by the comparison of their RT and MS
2 fragments with previous studies
[39][42][58,61]. Compound
19, m/z 505 [M − H]
−, was identified by GNPS as Quercetin-3-O-glucose-6″-acetate, while its isomer compound
21 was generically assigned as quercetin-O-acetyl hexoside
[41][60].
Compound
33, m/z 639 [M − H]
−, showed fragments at m/z 463 and 301 that correspond to the loss of glucuronyl and hexosyl moieties, respectively. Thus, on this evidence and coherently with a previous study
[43][62], we assigned this compound as quercetin-O-glucuronide-O-hexoside.
2.5. Identification of Isorhamnetin Derivatives
Compounds
28 and
34 showed a fragment ion at m/z 315 that is characteristic of the isorhamnetin aglycone after the NL of a glucuronyl [M − H-176]
− and hexose [M − H-162]
− moiety, respectively. Coherently with the literature, we assigned compound
28 as isorhamnetin3(7)-O-glucuronopyranoside and compound
34 as isorhamnetin-O-glucoside
[44][63].
2.6. Other Flavonoids Assignment
Compounds 5, 15, 16, 32, 35, and 36 were identified by GNPS as myricetin-3-O-β-D-galactopyranoside, astragaloside, scoparin, 6-O-methylscutellarin, 4-(3,4-Dihydroxyphenyl)-5-beta-D-glucopyranosyloxy-7-methoxycoumarin, and isookanin-7-O-glucoside, respectively.
We could not assign any identification to compounds 3, 7, 12, and 27.
2.7. DGMG Assignment
Compound 37 was assigned as digalactosylmonoacylglycerol (DGMG) 18:3 by GNPS annotation tool. By comparison, compounds 38 and 39 were tentatively assigned as DGMG 20:5 and DGMG 16:2, respectively.
2.8. Antiviral Activity
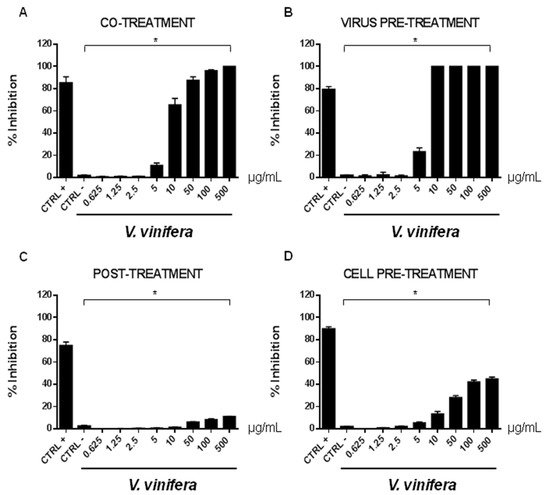
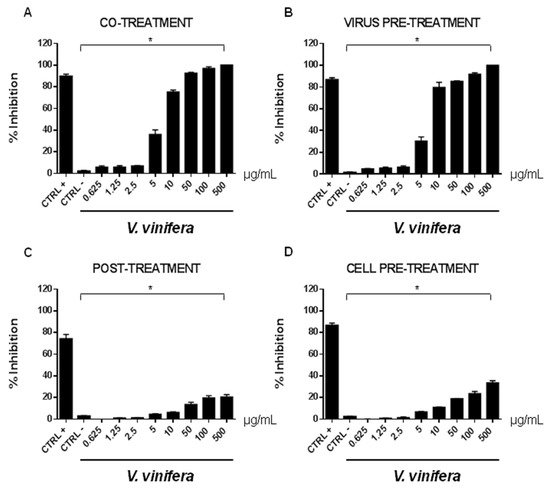
In order to evaluate if leaf polyphenol extract had an antiviral potential, and in which stage of the viral infection it could act, we performed experiments in four conditions: (a) simultaneous addition of extract and virus to the cells (“co-treatment”); (b) extract firstly incubated with the virus and then added to the cells (“virus pre-treatment”); (c) extract added to the infected cells (“post-treatment”); and (d) cells treated before with the extract and then infected with the virus (“cell pre-treatment”). The tested range of extract concentrations was from 0.625 to 500 µg/mL both for HSV-1 and SARS-CoV-2, which was used as a virus model for enveloped DNA and RNA viruses, respectively. The percentage of viral inhibition was calculated with respect to the untreated infected control (CTRL−). The reported antiviral activity was very similar for both viruses (Figure 3 and Figure 4), indicating an early effect manifesting directly on the viral envelope. The extract was most effective when incubated with HSV-1 and SARS-CoV-2 upon addition to target cells (co-treatment, Figure 3A and Figure 4A) or with the virus (virus pre-treatment, Figure 3B and Figure 4B) prior to infection on the cell monolayer. No activity was registered when the extract was added after HSV-1 infection (post-treatment, Figure 3C and Figure 4C), while a slight antiviral effect was observed when cells were firstly treated with the extract and then infected (cell pre-treatment, Figure 3D and Figure 4D). In detail, leaf extract was able to completely inhibit HSV-1 replication in virus pre-treatment assay until a concentration of 10 µg/mL was obtained but the virus resumed growing at 5 µg/mL, though with less efficiency (Figure 3B). Instead, the antiviral activity of V. vinifera extract was slightly lower against SARS-CoV-2, as it was set at 80% at 10 µg/mL. As HSV-1, no relevant inhibition was detected when the concentration of the extract was reduced to 5 µg/mL.
Figure 3. Antiviral effect (expressed as % of inhibition) of leaf extract (μg/mL) against HSV-1 in different plaque reduction assays: (A) co-treatment; (B) virus pre-treatment; (C) post-treatment; (D) cell pre-treatment. The results presented were obtained from three independent experiments. Data are mean ± SD. Statistical differences were analyzed via Student’s t-test, a value of p ≤ 0.05 was considered significant, with * p ≤ 0.05.
Figure 4. Antiviral effect (expressed as % of inhibition) of leaf extract (μg/mL) against SARS-CoV-2 in different plaque reduction assays: (A) co-treatment; (B) virus pre-treatment; (C) post-treatment; (D) cell pre-treatment. The results presented were obtained from three independent experiments. Data are mean ± SD. Statistical differences were analyzed via Student’s t-test, a value of p ≤ 0.05 was considered significant, with * p ≤ 0.05.
The leaf extract definitely showed a stronger antiviral potential than Greco extract, which was used as a positive control (CTRL+) against HSV-1 and SARS-CoV-2
[11][45][46][47][11,64,65,66].
These data are very interesting and indicate that the leaf extract could act directly on viral particles, blocking the interaction with the cell membrane. To deepen the viral inactivation mechanism, another antiviral test was carried out, such as the attachment assay (Figure 5 and Figure 6), by shifting the temperature during the infection at 4 °C.
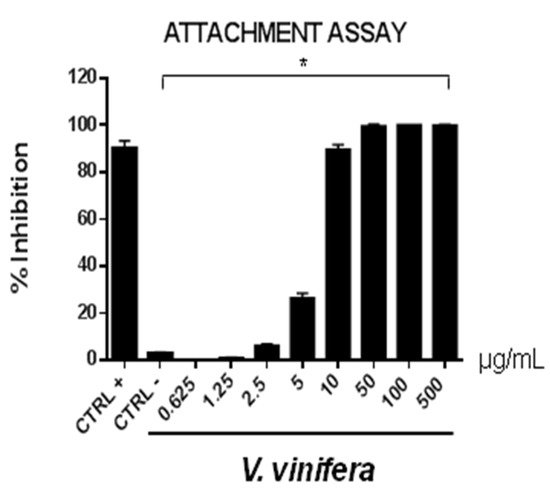
Figure 5. Early viral attachment assay of leaf extract (μg/mL) against HSV-1. The antiviral activity is expressed as % of inhibition. The results presented were obtained from three independent experiments. Data are mean ± SD. Statistical differences were analyzed via Student’s t-test, a value of p ≤ 0.05 was considered significant, with * p ≤ 0.05.
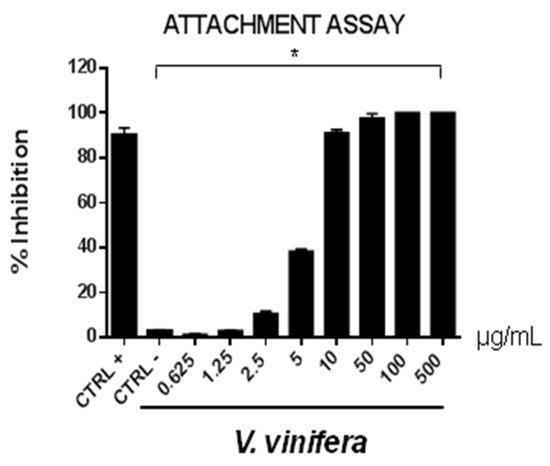
Figure 6. Early viral attachment assay of leaf extract (μg/mL) against SARS-CoV-2. The antiviral activity is expressed as % of inhibition. The results presented were obtained from three independent experiments. Data are mean ± SD. Statistical differences were analyzed via Student’s t-test, a value of p ≤ 0.05 was considered significant, with * p ≤ 0.05.
The extract’s inhibitory effects on virus-cell binding event revealed that it was able to block the attachment of both of the viruses into the host cell. It is most likely that leaf extract prevents the binding of viral envelope with the cell membrane and all the subsequent stages of infection by hindering some interacting site inside the viral glycoprotein deputed to the fusion, i.e., HSV-1 glycoprotein B and SARS-CoV-2 spike protein.
To validate data obtained by plaque assays, we analyzed the V. vinifera effect on expression genes involved in the viral infection (Figure 7 and Figure 8).
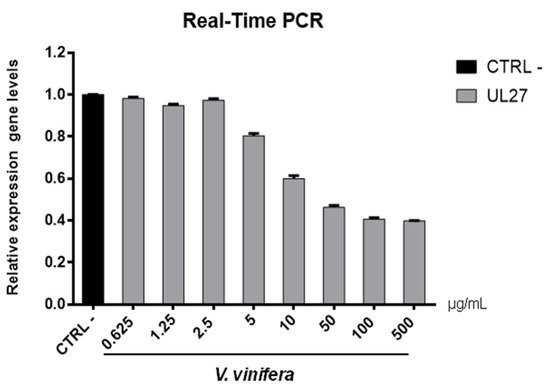
Figure 7. UL27 (glycoprotein B) gene expression analysis in Vero cells. V.vinifera leaf extract at 0.625, 1.25, 2.5, 5, 10, 50, 100, and 500 μg/mL was incubated with HSV-1 for 1 h and then, the mixture was added on Vero cells for another 1 h. After 2 days post-infection, RNA was extracted and Real-Time PCR was performed. Relative expression gene level was calculated with respect to CTRL−.
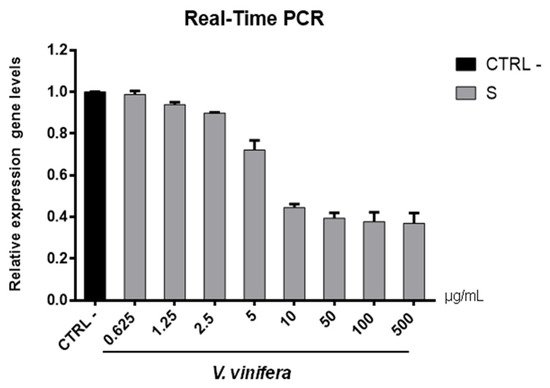
Figure 8. S gene expression analysis in Vero cells. V.vinifera leaf extract at 0.625, 1.25, 2.5, 5, 10, 50, 100, and 500 μg/mL was incubated with SARS-CoV-2 for 1 h and then the mixture was added on Vero cells for another 1 h. After 2 days post-infection, RNA was extracted, and Real-Time PCR was performed. Relative expression gene levels were calculated with respect to CTRL−.
Regarding the HSV-1 antiviral effect, UL27 gene, a late gene coding for the structural glycoprotein B (gB) was quantified via molecular assay (
Figure 7). In brief, a virus pre-treatment assay was performed, as previously mentioned, and RNA was collected after 48 h post-infection. Results showed that the extract inhibited HSV-1 replication by blocking much of viral gene expression, which almost halved at a concentration of 10 µg/mL. The outcomes on the replication are in accordance with literature
[48][49][50][51][52][67,68,69,70,71], demonstrating that the leaf extract had a virucidal action and could be used as a prophylactic treatment for herpetic infections. Anti-SARS-CoV-2 activity of V. vinifera leaf extract was also investigated through the spike protein (S) gene expression (
Figure 8). S protein represents the anti-receptor inserted into the viral envelope essential to the attachment to the host cell via the human angiotensin-converting enzyme 2 (hACE2) receptor
[53][54][55][72,73,74].
V. vinifera extract strongly reduced the S expression after SARS-CoV-2 infection until a concentration of 10 µg/mL was reached, and it resumed being expressed in a dose-dependent manner at lower concentrations. All together, these data indicated that V. vinifera leaf extract possesses a great antiviral and never previously investigated activity, which was reported against SARS-CoV-2. To date, the activity of flavonoids against SARS-CoV-2 has been analyzed with promising results in silico through bioinformatics approaches
[56][57][58][46,48,75]; in addition, the activity of these molecules towards other coronaviruses, such as the Severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS), has been widely reported
[59][76]. Altogether these results are noteworthy, although some phenolic compounds identified in the leaf extract are already reported as antiviral agents, such as quercetin which is currently under clinical trial
[60][77]. This study focuses on the high anti-inflammatory ability of quercetin, effective in COVID-19 treatment, and its dosage depends on the type of treatment: 500 mg for prophylactic use and 1000 mg for curative treatment.
2.9. Cytotoxicity Evaluation
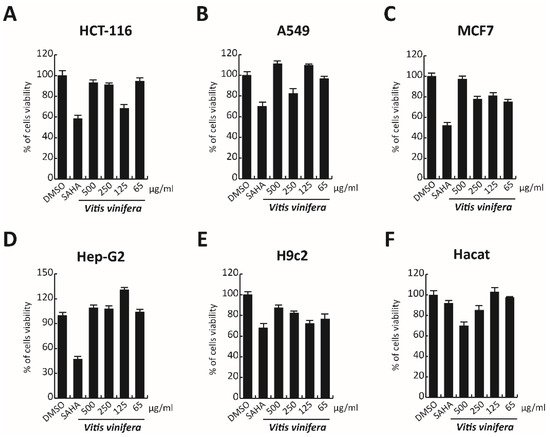
In order to complete the toxicological activity of leaf extract, V. vinifera was tested in colon, lung, breast, cardiomyocytes, hepatocyte cancer cells, and immortalized human keratinocyte cell line for 48 h at four different concentrations from 65 μg/mL to 500 μg/mL (
Figure 9A–F). With respect to the known published anticancer effect of the grape, seeds, or peel of V. vinifera
[61][78], the leaf extract did not induce any substantial variation in cell cancer viability. This different response, compared to that previously reported
[62][79], could be explained by the different extraction methodologies or V. vinifera varieties used. These data suggest how this complex polyphenolic extract, in these conditions, was able to selectively act on a specific target, without altering other cellular mechanisms.
Figure 9. (A–F) Cell viability evaluation by MTT assay of HCT-116, A549, MCF7, cardiomyocytes, Hep-G2, and Hacat cells after treatment with V. vinifera for 48 h at 500, 250, 125, and 65 μg/mL. SAHA was used as a control at 5 μM for 24 h.