1. Overview
Enhancers are critical genomic elements that can cooperate with promoters to regulate gene transcription in both normal and cancer cells. Recent studies reveal that enhancer regions are transcribed to produce a class of noncoding RNAs referred to as enhancer RNAs (eRNAs). Emerging evidence shows that eRNAs play important roles in enhancer activation and enhancer-driven gene regulation, and the expression of eRNAs may be a critical factor in tumorigenesis. The important roles of eRNAs in cancer signaling pathways are also gradually unveiled, providing a new insight into cancer therapy. Here, we review the roles of eRNAs in regulating cancer signaling pathways and discuss the potential of eRNA-targeted therapy for human cancers.
2. Enhancers
Enhancers are regions of DNA that drive transcription independently of their distance and orientation from the gene, in contrast to promoters which exert their functions in an orientation-dependent manner. Enhancers contain binding sites for RNA polymerase (RNA pol) II, multiple transcription factors, and co-regulators, and can interact with promoters to form spatial chromatin loops. As critical genomic elements, enhancers regulate gene transcription in various diseases including human cancers, and enhancer malfunctions can have a direct effect on tumor growth.
Enhancer regions themselves can also be actively transcribed and produce a type of non-coding RNA termed as enhancer RNAs (eRNAs), which were first detected in 2010
[1]. eRNAs are nascent RNA transcripts that exhibit a 5′ cap, and they were first observed to be short bidirectionally transcribed RNA transcripts of no more than 2 kb and were non-polyadenylated
[1]. Soon after, it was discovered that there were also polyadenylated eRNAs, and interestingly, these polyadenylated eRNAs were generally unidirectionally transcribed and longer than those that were non-polyadenylated. However, according to FANTOM5 enhancer atlas
[2], bidirectionally transcribed, non-polyadenylated eRNAs are more common, and the majority of eRNAs are not spliced. The process of eRNA transcription is triggered by the binding of specific transcription factors and coactivators to enhancers. Then, they recruit other transcription factors, complex and histone modifiers such as P300/CBP. H3K27 is acetylated by P300/CBP and the enhancer regions are further opened, leading to the recruitment of essential proteins such as RNA pol II. Cofactors such as BRD4 promote RNA pol II elongation and eRNA processing
[3]. Although eRNAs are commonly observed, their roles are largely unknown, and it has been suggested that they are a byproduct of transcription. However, a growing number of studies have indicated diverse roles for eRNAs in regulating gene transcription and many other aspects of cell functions. Here, we review the roles of eRNAs in cancer gene expression and signaling pathways and discuss the potential of eRNA-targeted therapy for human cancers.
eRNAs define active enhancers and co-localize with epigenetic markers, and it is widely accepted that eRNA production is closely associated with enhancer activation. Indeed, the expression of eRNAs positively correlates with the level of histone H3 acetylation on lysine 27 (H3K27ac), which is a marker for active enhancers
[4]. Moreover, eRNA transcription is reported to be a better marker than H3K27ac for identifying enhancer activation and is shown to forecast novel enhancers
[5][6][7][5,6,7].
Super enhancers (SEs) consist of large clusters of transcriptional enhancers that activate cell type- and tissue-specific genes. Compared with typical enhancers, SEs have specific features such as a significantly increased transcription factor occupancy, high levels of H3K4me1 and H3K27ac density, and increased occupancy of RNA pol II, mediator, p300/CBP, and bromodomain-containing (BRD)4 cofactors. SE transcription also produces higher quantities of eRNAs
[8][9][10][11][12][8,9,10,11,12].
eRNA transcription via RNA pol II precedes the activation of the target gene, suggesting that eRNAs are regulators of enhancer-mediated target gene expression
[7][13][14][15][16][7,13,14,15,16]. Indeed, the production of eRNAs is positively correlated with the expression of the target genes of corresponding enhancers
[1][2][3][17][1,2,3,17], with eRNA knockdown typically reducing target gene expression. For example, eRNA production from p53-bound enhancer regions (p53BERs) is required for the p53-dependent activation of gene expression, while the knockdown of p53BER2- and p53BER4-derived eRNAs inhibit the induction of genes closest to these two p53BER domains
[18].
eRNAs appear to play key roles in binding transcription factors needed for the up-regulation of enhancer-mediated genes; for example, colon cancer-associated transcript (CCAT) 1 recruited transcription factors TP63 and SOX2 to SE regions of the epidermal growth factor receptor gene (EGFR) to promote its transcription
[19]. Similarly, interactions have been reported between eRNAs and specific transcription factors, such as BRD4, P53, and cohesin, which bind a broad array of enhancers to promote gene transcription
[11][15][20][11,15,20]. Although enhancers are cis-acting regulatory elements located some distance from transcription start sites, they still effectively promote the expression of target genes independently of their position and orientation because they can loop over long genomic ranges to engage distant promoters. Previous studies suggested that eRNAs might promote enhancer–promoter looping and recruit transcription factors to specific enhancers
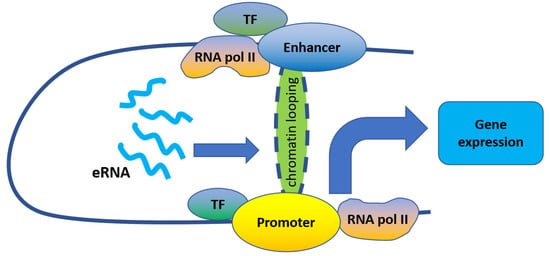
[8][21][22][23][8,21,22,23] ().
Figure 1. eRNAs regulate gene expression by promoting enhancer–promoter looping.
Indeed, eRNAs were reported to act mostly in-cis to promote enhancer–promoter looping. For example, eRNAs transcribed from enhancers adjacent to 17β-estradiol up-regulated genes play important roles in their induction by strengthening enhancer–promoter looping through estrogen receptor α binding
[20]. Similarly, eRNAs produced from an enhancer located upstream of the kallikrein-related peptidase 3 gene (KLK3) promote the interaction of the KLK3 enhancer and KLK2 promoter, thus enhancing the long-distance transcriptional activation of KLK2
[13]. Furthermore, Epstein Barr virus (EBV) SE eRNAs are important for enhancer–promoter looping at the MYC locus, while eRNA CCAT1 interacts with CTCF to promote chromatin looping between the MYC promoter and enhancers, leading to MYC gene expression
[24]. As such, eRNA knockdown can disrupt the loop structures and decrease target gene transcription
[13][24][25][26][13,24,25,26].
3. eRNA in Oncogene Expression Regulation and Tumorigenesis
Although the eRNA molecular mechanisms in human cancers are not yet fully understood, abundant evidence suggests that the abnormal expression of eRNAs is closely related to tumorigenesis. Zhang et al. examined differentially expressed eRNAs in matched tumor-normal samples across 16 cancer types, and found more up-regulated eRNAs (median, 42.2%) than down-regulated ones (median, 9.9%)
[27]. Similarly, Qin et al. observed elevated global eRNA expression among lung adenocarcinoma tumor samples compared with control samples
[28]. It has also been reported that individual eRNAs play important roles in tumorigenesis in many cancer types (including lung adenocarcinoma, prostate cancer, breast cancer, and squamous cell carcinomas), mainly through regulating carcinoma-related genes.
In lung adenocarcinoma, Qin et al. identified hundreds of eRNAs to be functional as their correlated genes were over-represented in cancer driver and clinically actionable gene ensemble. The eRNA located upstream of telomerase reverse transcriptase (TERT), a well-known predisposition gene for lung cancer
[29], appears to contribute to cancer development by up-regulating TERT expression. Moreover, the copy number amplification of Forkhead box (FOX)O6 eRNA contributes to the up-regulation of FOXO6 in lung adenocarcinoma
[28].
Similarly, a group of androgen receptor (AR)-regulated eRNAs, including prostate-specific antigen eRNA (PSA eRNA), were found to be up-regulated in castration-resistant prostate cancer (CRPC) cells, patient-derived xenografts, and patient tissues. These AR-regulated eRNAs have important roles in the regulation of AR target genes. For example, PSA eRNA bound cyclin T1 and activated the positive transcription elongation factor, then increased serine-2 phosphorylation of RNA pol II (Pol II-Ser2p), which is essential for transcription elongation and gene expression. Additionally, PSA eRNA knockdown significantly decreased Pol II-Ser2p levels at the loci of a subset of genes, including those involved in regulating the cell cycle, growth, survival, migration, and invasion, such as VEGFA, NCAPD3, ADAMTS1, and IGF1R. An HIV-1 trans-activation response element RNA-like motif in PSA eRNA was also shown to be essential for increased Pol II-Ser2p occupancy levels and CRPC cell growth
[30].
The high expression of NET1e, an eRNA located about 90 kb downstream of the oncogene NET1, was detected in breast cancer, and NET1e knockdown significantly reduced the proliferation of the breast cancer cell line MCF7. NET1e appears to contribute to breast cancer progression by up-regulating NET1 expression
[27].
Finally, the knockdown of oncogenic eRNA CCAT1 in squamous cell carcinomas cells down-regulates the expression of SE-associated genes involved in cell proliferation, growth, and migration, as well as those with a role in DNA-protein interactions. This suggests that CCAT1 functions in cancer progression and the interaction of macromolecules in cancer
[19].
Taken together, these results suggest important roles for eRNAs in various cancer types.
4. Conclusions
Identifying the critical role of eRNAs in regulating gene transcription and in the development of different cancer types has also enabled a better understanding of enhancers. Recent studies suggest that eRNAs regulate gene expression through promoting enhancer–promoter looping, and the knockdown of eRNAs is often accompanied by decreased target gene expression. Abundant evidence shows that eRNAs are abnormally expressed in various cancers and that their expression is closely related to tumorigenesis.
The expression of genes in signaling pathways correlates with eRNAs in various types of cancer. Thus, eRNA expression is regulated by signaling pathways, and, in turn, eRNAs also regulate certain signaling pathways. Although the associations between eRNAs and signaling pathways in human cancers are well-studied, the underlying molecular mechanisms involved remain unclear. Additionally, the relationship between eRNAs and signaling pathways has been mainly established through genomic research, with limited supporting evidence from molecular assays. Therefore, further molecular analyses are required to fully understand these mechanisms.
The characterization of eRNA functional roles in cancers may provide novel insights into cancer therapy. For example, the knockdown of specific eRNAs inhibits cell proliferation, invasion, and migration in cancer; thus, eRNA-targeted inhibitors could provide new entry points for cancer treatments. Additionally, the potential clinical value of eRNAs is suggested by the establishment of a correlation between eRNAs and clinically actionable genes and immune checkpoints. However, because cases that directly apply eRNAs to cancer treatment are limited, further efforts are required to fully explore the utility of eRNAs in cancer therapy.
At present, several eRNA data resources have been established, providing the research community with convenient and efficient platforms that facilitate the exploration for functional roles of eRNAs in both normal cells and cancer cells. With the boom of research on eRNA in the future and the growing number of samples coming along, these eRNA data resources may need to be updated continuously.