Placental disorders are a major cause of pregnancy loss in humans, and 40%–60% of embryos are lost between fertilization and birth. Successful embryo implantation and placental development requires rapid proliferation, invasion, and migration of trophoblast cells. In recent years, microRNAs (miRNAs) have emerged as key regulators of molecular pathways involved in trophoblast function. A miRNA binds its target mRNA in the 3ʹ-untranslated region (3ʹ-UTR), causing its degradation or translational repression. Lethal-7 (
let-7
) miRNAs induce cell differentiation and reduce cell proliferation by targeting proliferation-associated genes. The oncoprotein LIN28 represses the biogenesis of mature
let-7
miRNAs. Proliferating cells have high LIN28 and low
let-7
miRNAs, whereas differentiating cells have low LIN28 and high
let-7
miRNAs. In placenta, low LIN28 and high
let-7
miRNAs can lead to reduced proliferation of trophoblast cells, resulting in abnormal placental development. In trophoblast cells,
let-7
miRNAs reduce the expression of proliferation factors either directly by binding their mRNA in 3ʹ-UTR or indirectly by targeting the -rich interaction domain (ARID)3B complex, a transcription-activating complex comprised of ARID3A, ARID3B, and histone demethylase 4C (KDM4C). In this review, we discuss regulation of trophoblast function by miRNAs, focusing on the role of LIN28-
let-7-
ARID3B pathway in placental development.
- miRNA
- trophoblast cells
- cell proliferation
- ARID3B complex
1. LIN28-let-7-ARID3B Pathway in Trophoblast Cells
LIN28-let-7-ARID3B Pathway in Trophoblast Cells
AT-rich interactive domain (ARID) proteins, first recognized in 1997, are a family of 15 proteins which binds to AT-rich regions of DNA
. ARID proteins play an important role in cell proliferation, differentiation, and development, and are upregulated in tumorous tissues
[3]
. The subfamilies of ARID proteins include ARID1, ARID2, ARID3, ARID4, ARID5, JARID1, and JARID2. The ARID3 subfamily has three members including ARID3A, ARID3B, and ARID3C. Although most of the ARID proteins act as tumor suppressors, ARID3A and ARID3B promote tumorigenesis
[1]
. ARID3A inhibits cell differentiation, promotes cell proliferation, and increases survival potential of cells
, whereas ARID3B promotes proliferation, invasion, and migration of cancer cells
. However, ARID3B is more widely expressed in different tissues compared to ARID3A, suggesting more involvement of ARID3B in biological functions
[2]
. ARID3A and ARID3B are structurally similar and bind a similar region of DNA. Both ARID3A and ARID3B have an extended central ARID domain and two conserved amino acid domains at the C-terminal, termed REKLES α and REKLES β. Only members of the ARID3 subfamily have REKLES domains
[8]
.
In contrast to ARID3B which is exclusively localized in the nucleus, ARID3A shuttles between the nucleus and cytoplasm. Once in the nucleus, ARID3A interacts with ARID3B through REKLES β domain. Therefore localization of ARID3A in the nucleus is dependent on its interaction with ARID3B in the nucleus, suggesting the dominant role of ARID3B
. In cancer cells, ARID3A and ARID3B recruit histone demethylase 4C (KDM4C) to make a tri-protein complex, called the ARID3B complex
[10]
. The ARID3B complex binds in the promoter areas of stemness genes and
let-7
target genes, leading to histone demethylation by KDM4C and increased gene expression by initiation of transcription
[10]
. Therefore, genes regulated by the ARID3B complex also include
let-7
miRNA target genes. Liao et al. further demonstrated that both ARID3A and ARID3B are targeted by
let-7
miRNAs. Hence, other than directly targeting the mRNAs of target genes,
let-7
miRNA can indirectly regulate their target genes by targeting and reducing the expression of
ARID3A
and
ARID3B (Figure 1)
(Figure 4)
.
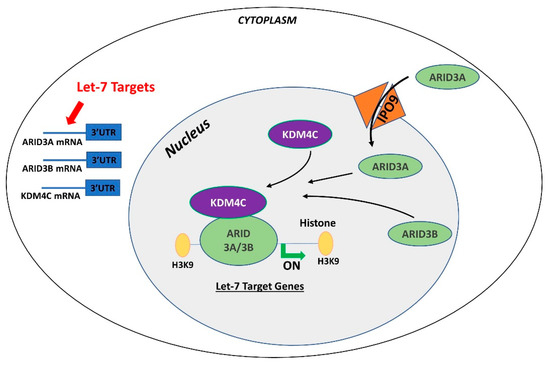
Figure 1. Gene regulation by the AT-rich interactive domain (ARID)3B complex. ARID3A is imported in the nucleus by importin 9 (IPO9), where it binds ARID3B and histone demethylase 4C (KDM4C) to form the ARID3B complex. The ARID3B complex binds in the promoter regions and activates transcription of let-7 target genes. Other than directly binding the mRNA of their target genes, let-7 miRNAs also target the ARID3B complex and reduce its expression, ultimately leading to reduced expression of let-7 target genes
Both ARID3A and ARID3B have high expression in human trophoblast cells
Figure 4. Gene regulation by the AT-rich interactive domain (ARID)3B complex. ARID3A is imported in the nucleus by importin 9 (IPO9), where it binds ARID3B and histone demethylase 4C (KDM4C) to form the ARID3B complex. The ARID3B complex binds in the promoter regions and activates transcription of let-7 target genes. Other than directly binding the mRNA of their target genes, let-7 miRNAs also target the ARID3B complex and reduce its expression, ultimately leading to reduced expression of let-7 target genes.
Both ARID3A and ARID3B have high expression in human trophoblast cells
ARID3A knockout mice have severe structural defects in placenta
[14]
. In ACH-3P cells, ARID3A, ARID3B, and KDM4C make the tri-protein ARID3B complex
[11]
. In term human placentas from IUGR pregnancies, LIN28A and LIN28B are low,
let-7
miRNAs are high, and ARID3A and ARID3B are low, which suggest a correlation between LIN28-
let-7
miRNA axis and the ARID3B complex
[11]
. Due to the well-established pathway of regulation of
let-7
target genes through ARID3B complex and their role in cell proliferation, it is important to understand this phenomenon in early placental development. We recently showed the correlation between LIN28-
let-7
miRNA axis and the ARID3B complex using ACH-3P and Sw.71 cells. Double knockout of LIN28A and LIN28B in ACH-3P cells increases
let-7
miRNAs and decreases the expression ARID3A, ARID3B, and KDM4C. Similarly, double knockin of LIN28A and LIN28B in Sw.71 cells decreases
let-7
miRNAs and increases expression of ARID3A, ARID3B, and KDM4C
[11]
. In trophoblast cells, the ARID3B complex binds to the promoter areas of proliferation-associated
let-7
target genes including
HMGA1, c-MYC, VEGF-A
, and
WNT1,
facilitating their transcription via KDM4C mediated histone demethylation. ARID3B knockout ACH-3P cells and KDM4C cannot be recruited in the promoter regions of
HMGA1, c-MYC, VEGF-A
and
WNT1
, and expression of these genes is also significantly reduced
[15]
. Moreover, ARID3B knockout ACH-3P cells have a reduced proliferation rate compared to control cells
[11]
. Knockdown of LIN28A or LIN28B in sheep trophectoderm increases
let-7
miRNAs and reduces the expression of ARID3A and ARID3B
[16]
, showing regulation of the ARID3B complex by LIN28-
let-7
miRNA axis in vivo. Collectively these findings show that
let-7
miRNAs target the ARID3B complex in trophoblast cells, and the ARID3B complex regulates genes with known importance in placental development.
2. Conclusions
Conclusions
For a long time, transcription activating proteins were thought to be the main regulators of gene expression in cells. However, in recent years microRNAs have emerged as “regulators of the regulators”. miRNAs regulate important processes in trophoblast cells including cell proliferation, differentiation, invasion, and migration. Although identification of widespread let-7 miRNA target genes makes them an important player in placental development, the role of other miRNAs and molecular pathways in placental development cannot be ignored. Although different studies have reported reduced LIN28 and high let-7 miRNAs in term human placentas from preeclamptic and IUGR pregnancies, the exact cause of this dysregulation is not clear. Epigenetic modifications due to adverse uterine environment, random genetic mutations, prenatal insults like hypoxia, oxidative stress, maternal malnutrition, and gestational stress are some of the possible factors which can dysregulate LIN28-let-7 axis in trophoblast cells. LIN28-let-7-ARID3B pathway regulates trophoblast cell proliferation by modulating the expression of proliferation associated genes in vitro and in vivo. The trophoblast cells with low LIN28 will have high let-7 miRNAs and low ARID3B. There are two possible pathways of regulation of proliferation-associated genes by let-7 miRNAs in trophoblast cells (Figure 2). One pathway involves binding of let-7 miRNAs in 3ʹ-UTR of their target mRNA leading to mRNA degradation or translational repression. Secondly, let-7 miRNAs target ARID3A, ARID3B, and KDM4C to inhibit or reduce transcriptional activation of proliferation-associated genes by the ARID3B complex. Therefore, trophoblast cells with high let-7 miRNAs will have reduced expression of proliferation factors and more of a tendency to differentiate. High let-7 miRNAs during early placental development reduce cell proliferation, invasion, and migration of trophoblast cells, leading to placental abnormalities. Let-7 miRNAs might be the major players in pathogenesis of placenta-associated disorders. miRNAs can be readily measured in peripheral blood, tissue biopsies, saliva, cerebrospinal fluid, urine, and other biological samples. High let-7 miRNAs are upregulated in IUGR and preeclamptic placentas, suggesting that let-7 miRNAs can be potential biomarkers for early diagnosis of PE and IUGR. It remains to be explored if early stage placentas from compromised pregnancies will have high let-7 miRNAs and will the increase in let-7 miRNAs in placenta be reflected in maternal blood. Animal models of IUGR can be used to answer these questions.
For a long time, transcription activating proteins were thought to be the main regulators of gene expression in cells. However, in recent years microRNAs have emerged as “regulators of the regulators”. miRNAs regulate important processes in trophoblast cells including cell proliferation, differentiation, invasion, and migration. Although identification of widespread let-7 miRNA target genes makes them an important player in placental development, the role of other miRNAs and molecular pathways in placental development cannot be ignored. Although different studies have reported reduced LIN28 and high let-7 miRNAs in term human placentas from preeclamptic and IUGR pregnancies, the exact cause of this dysregulation is not clear. Epigenetic modifications due to adverse uterine environment, random genetic mutations, prenatal insults like hypoxia, oxidative stress, maternal malnutrition, and gestational stress are some of the possible factors which can dysregulate LIN28-let-7 axis in trophoblast cells. LIN28-let-7-ARID3B pathway regulates trophoblast cell proliferation by modulating the expression of proliferation associated genes in vitro and in vivo. The trophoblast cells with low LIN28 will have high let-7 miRNAs and low ARID3B. There are two possible pathways of regulation of proliferation-associated genes by let-7 miRNAs in trophoblast cells (Figure 5). One pathway involves binding of let-7 miRNAs in 3ʹ-UTR of their target mRNA leading to mRNA degradation or translational repression. Secondly, let-7 miRNAs target ARID3A, ARID3B, and KDM4C to inhibit or reduce transcriptional activation of proliferation-associated genes by the ARID3B complex. Therefore, trophoblast cells with high let-7 miRNAs will have reduced expression of proliferation factors and more of a tendency to differentiate. High let-7 miRNAs during early placental development reduce cell proliferation, invasion, and migration of trophoblast cells, leading to placental abnormalities. Let-7 miRNAs might be the major players in pathogenesis of placenta-associated disorders. miRNAs can be readily measured in peripheral blood, tissue biopsies, saliva, cerebrospinal fluid, urine, and other biological samples. High let-7 miRNAs are upregulated in IUGR and preeclamptic placentas, suggesting that let-7 miRNAs can be potential biomarkers for early diagnosis of PE and IUGR. It remains to be explored if early stage placentas from compromised pregnancies will have high let-7 miRNAs and will the increase in let-7 miRNAs in placenta be reflected in maternal blood. Animal models of IUGR can be used to answer these questions.
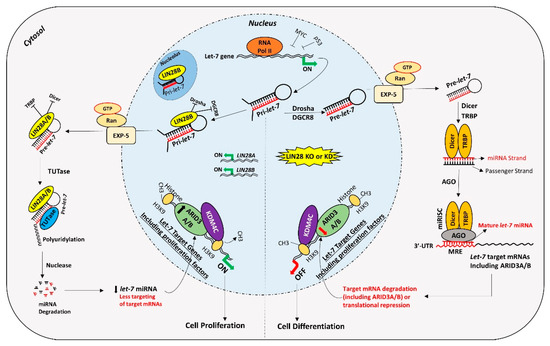
Figure 2. Proposed mechanism for gene regulation in trophoblast cells. Left panel of figure: LIN28 represses the biogenesis of mature let-7 miRNAs by binding pri-let-7 and pre-let-7 miRNAs and inhibiting their processing. Due to the low level of mature let-7 miRNAs in the cells, there will be less targeting of proliferation-associated genes. Moreover, the ARID3B complex will initiate the transcription of proliferation-associated genes and increase their expression. Increased expression of proliferation-associated genes will lead to increased cell proliferation. Right panel of figure: If LIN28 is knocked-out or knocked-down, there will be no suppression of let-7 miRNA biogenesis. High let-7 miRNAs will target and reduce the expression of proliferation associated genes and the ARDI3B complex, driving the cells towards differentiation.
Figure 5. Proposed mechanism for gene regulation in trophoblast cells. Left panel of figure: LIN28 represses the biogenesis of mature let-7 miRNAs by binding pri-let-7 and pre-let-7 miRNAs and inhibiting their processing. Due to the low level of mature let-7 miRNAs in the cells, there will be less targeting of proliferation-associated genes. Moreover, the ARID3B complex will initiate the transcription of proliferation-associated genes and increase their expression. Increased expression of proliferation-associated genes will lead to increased cell proliferation. Right panel of figure: If LIN28 is knocked-out or knocked-down, there will be no suppression of let-7 miRNA biogenesis. High let-7 miRNAs will target and reduce the expression of proliferation associated genes and the ARDI3B complex, driving the cells towards differentiation.
References
- Wilsker, D.; Patsialou, A.; Dallas, P.; Moran, E. ARID proteins:A diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. Mol. Biol. J. Am. Assoc. Cancer Res. 2002, 13, 95–106.Wilsker, D.; Patsialou, A.; Dallas, P.; Moran, E. ARID proteins:A diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. Mol. Biol. J. Am. Assoc. Cancer Res. 2002, 13, 95–106. [Google Scholar]
- Hong, Z.; Wei, S.; Bi, X.; Zhao, J.; Huang, Z.; Li, Z.; Zhou, J.; Cai, J.; Chen, L.; Lin, C.; et al. Recent advances in the ARID family:Focusing on roles in human cancer. OncoTargets Ther. 2014, 7, 315–324.Hong, Z.; Wei, S.; Bi, X.; Zhao, J.; Huang, Z.; Li, Z.; Zhou, J.; Cai, J.; Chen, L.; Lin, C.; et al. Recent advances in the ARID family:Focusing on roles in human cancer. OncoTargets Ther. 2014, 7, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Fukuyo, Y.; Takahashi, A.; Hara, E.; Horikoshi, N.; Pandita, T.K.; Nakajima, T. E2FBP1 antagonizes the p16INK4A-Rb tumor suppressor machinery for growth suppression and cellular senescence by regulating promyelocytic leukemia protein stability. Int. J. Oral Sci. 2011, 3, 200–208.Fukuyo, Y.; Takahashi, A.; Hara, E.; Horikoshi, N.; Pandita, T.K.; Nakajima, T. E2FBP1 antagonizes the p16INK4A-Rb tumor suppressor machinery for growth suppression and cellular senescence by regulating promyelocytic leukemia protein stability. Int. J. Oral Sci. 2011, 3, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Fukuyo, Y.; Mogi, K.; Tsunematsu, Y.; Nakajima, T. E2FBP1/hDril1 modulates cell growth through downregulation of promyelocytic leukemia bodies. Cell Death Differ. 2004, 11, 747–759.Fukuyo, Y.; Mogi, K.; Tsunematsu, Y.; Nakajima, T. E2FBP1/hDril1 modulates cell growth through downregulation of promyelocytic leukemia bodies. Cell Death Differ. 2004, 11, 747–759. [Google Scholar] [CrossRef]
- Kobayashi, K.; Jakt, L.M.; Nishikawa, S.-I. Epigenetic regulation of the neuroblastoma genes, Arid3b and Mycn. Oncogene 2012, 32, 2640–2648.Kobayashi, K.; Jakt, L.M.; Nishikawa, S.-I. Epigenetic regulation of the neuroblastoma genes, Arid3b and Mycn. Oncogene 2012, 32, 2640–2648. [Google Scholar] [CrossRef]
- Bobbs, A.; Gellerman, K.; Hallas, W.M.; Joseph, S.; Yang, C.; Kurkewich, J.; Dahl, K.C. ARID3B Directly Regulates Ovarian Cancer Promoting Genes. PLoS ONE 2015, 10, e0131961.Bobbs, A.; Gellerman, K.; Hallas, W.M.; Joseph, S.; Yang, C.; Kurkewich, J.; Dahl, K.C. ARID3B Directly Regulates Ovarian Cancer Promoting Genes. PLoS ONE 2015, 10, e0131961. [Google Scholar] [CrossRef]
- Nakahara, S.; Fukushima, S.; Yamashita, J.; Kubo, Y.; Tokuzumi, A.; Miyashita, A.; Harada, M.; Nakamura, K.; Jinnin, M.; Ihn, H. AT-rich Interaction Domain-containing Protein 3B is a New Tumour Marker for Melanoma. Acta Derm. Venereol. 2017, 97, 112–114.Nakahara, S.; Fukushima, S.; Yamashita, J.; Kubo, Y.; Tokuzumi, A.; Miyashita, A.; Harada, M.; Nakamura, K.; Jinnin, M.; Ihn, H. AT-rich Interaction Domain-containing Protein 3B is a New Tumour Marker for Melanoma. Acta Derm. Venereol. 2017, 97, 112–114. [Google Scholar] [CrossRef]
- Kim, D.; Probst, L.; Das, C.; Tucker, P.W. REKLES Is an ARID3-restricted Multifunctional Domain. J. Biol. Chem. 2007, 282, 15768–15777.Kim, D.; Probst, L.; Das, C.; Tucker, P.W. REKLES Is an ARID3-restricted Multifunctional Domain. J. Biol. Chem. 2007, 282, 15768–15777. [Google Scholar] [CrossRef]
- Webb, C.F.; Bryant, J.; Popowski, M.; Allred, L.; Kim, D.; Harriss, J.; Schmidt, C.; Miner, C.A.; Rose, K.; Cheng, H.-L.; et al. The ARID Family Transcription Factor Bright Is Required for both Hematopoietic Stem Cell and B Lineage Development. Mol. Cell. Biol. 2011, 31, 1041–1053.Webb, C.F.; Bryant, J.; Popowski, M.; Allred, L.; Kim, D.; Harriss, J.; Schmidt, C.; Miner, C.A.; Rose, K.; Cheng, H.-L.; et al. The ARID Family Transcription Factor Bright Is Required for both Hematopoietic Stem Cell and B Lineage Development. Mol. Cell. Biol. 2011, 31, 1041–1053. [Google Scholar] [CrossRef]
- Liao, T.-T.; Hsu, W.-H.; Ho, C.-H.; Hwang, W.-L.; Lan, H.-Y.; Lo, T.; Chang, C.-C.; Tai, S.; Yang, M.-H. Let-7 Modulates Chromatin Configuration and Target Gene Repression through Regulation of the ARID3B Complex. Cell Rep. 2016, 14, 520–533.Liao, T.-T.; Hsu, W.-H.; Ho, C.-H.; Hwang, W.-L.; Lan, H.-Y.; Lo, T.; Chang, C.-C.; Tai, S.; Yang, M.-H. Let-7 Modulates Chromatin Configuration and Target Gene Repression through Regulation of the ARID3B Complex. Cell Rep. 2016, 14, 520–533. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Anthony, R.V.; Bouma, G.J.; Winger, Q.A. LIN28-let-7 axis regulates genes in immortalized human trophoblast cells by targeting the ARID3B-complex. FASEB J. 2019, 33, 12348–12363.Ali, A.; Anthony, R.V.; Bouma, G.J.; Winger, Q.A. LIN28-let-7 axis regulates genes in immortalized human trophoblast cells by targeting the ARID3B-complex. FASEB J. 2019, 33, 12348–12363. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250.Uhlén, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef] [PubMed]
- Rhee, C.; Lee, B.-K.; Beck, S.; Anjum, A.; Cook, K.R.; Popowski, M.; Tucker, H.O.; Kim, J. Arid3a is essential to execution of the first cell fate decision via direct embryonic and extraembryonic transcriptional regulation. Genome Res. 2014, 28, 2219–2232.Rhee, C.; Lee, B.-K.; Beck, S.; Anjum, A.; Cook, K.R.; Popowski, M.; Tucker, H.O.; Kim, J. Arid3a is essential to execution of the first cell fate decision via direct embryonic and extraembryonic transcriptional regulation. Genome Res. 2014, 28, 2219–2232. [Google Scholar] [CrossRef] [PubMed]
- Rhee, C.; Edwards, M.; Dang, C.; Harris, J.; Brown, M.; Kim, J.; Tucker, H.O. ARID3A is required for mammalian placenta development. Dev. Biol. 2016, 422, 83–91.Rhee, C.; Edwards, M.; Dang, C.; Harris, J.; Brown, M.; Kim, J.; Tucker, H.O. ARID3A is required for mammalian placenta development. Dev. Biol. 2016, 422, 83–91. [Google Scholar] [CrossRef]
- Brkić, J.; Dunk, C.; O’Brien, J.; Fu, G.; Nadeem, L.; Wang, Y.-L.; Rosman, D.; Salem, M.; Shynlova, O.; Yougbaré, I.; et al. MicroRNA-218-5p Promotes Endovascular Trophoblast Differentiation and Spiral Artery Remodeling. Mol. Ther. 2018, 26, 2189–2205.Brkić, J.; Dunk, C.; O’Brien, J.; Fu, G.; Nadeem, L.; Wang, Y.-L.; Rosman, D.; Salem, M.; Shynlova, O.; Yougbaré, I.; et al. MicroRNA-218-5p Promotes Endovascular Trophoblast Differentiation and Spiral Artery Remodeling. Mol. Ther. 2018, 26, 2189–2205. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Stenglein, M.D.; Spencer, T.E.; Bouma, G.J.; Anthony, R.V.; Winger, Q.A. Trophectoderm-Specific Knockdown of LIN28 Decreases Expression of Genes Necessary for Cell Proliferation and Reduces Elongation of Sheep Conceptus. Int. J. Mol. Sci. 2020, 21, 2549.Ali, A.; Stenglein, M.D.; Spencer, T.E.; Bouma, G.J.; Anthony, R.V.; Winger, Q.A. Trophectoderm-Specific Knockdown of LIN28 Decreases Expression of Genes Necessary for Cell Proliferation and Reduces Elongation of Sheep Conceptus. Int. J. Mol. Sci. 2020, 21, 2549. [Google Scholar] [CrossRef] [PubMed]
