1. Introduction
Wheat feeds one-quarter of the annual worldwide demand for plant proteins (60 metric tons) and has been the source of nutrition since the dawn of human civilization
[1][2][3][4]. It is not only a primary staple worldwide. But it is also responsible for numerous foodborne disorders, which remained one of the major causes of premature deaths in the most resource-deprived parts of the world since the prehistoric-times
[3][5][6][7][8]. Given its vast influence on human health, domestication of bread wheat and its subsequent industrialization has been considered a “mistake of evolution” that created conditions for human diseases related to gluten exposure
[7][9][10]. The word “gluten” refers to a complex mixture of proline and glutamine-rich seed-storage proteins that serves as fuel for multiple disorders
[11][12][13][14][15][16]. Different overlapping or non-overlapping epitopes have been shown to elicit various reactions in different individuals in accordance to their genetic constitutions
[14][17][18][19][20]. Gluten-intake in sensitive individuals can lead to gastrointestinal, neurological, and fatal symptoms such as non-Hodgkin lymphoma
[21][22][23]. These symptoms can be classified grossly into celiac disease, wheat sensitivity, and allergy
[11][23][24], with the celiac disease being the most prevalent gastrointestinal disorder
[23]. In celiac disease, the response to gluten is mediated by the adaptive immune system and the induction of autoantibodies against the indigestible gluten peptides and tissue transglutaminase 2 (tTG2), an enzyme involved in tissue homeostasis
[25]. The tTG2 is also responsible for chemical modification of gluten peptides, which enhance their recognition by the immune system. In this process, the faulty immune system in genetically predisposed individuals recognizes tTG2 as an enemy and triggers an autoimmune response against it
[14][26]. Because of their mode of action, gluten peptides were compared with the non-replicating pathogen
[27]. Since like pathogens, these peptides evade “host” defenses by escaping digestion through gastrointestinal enzymes, invade the intestinal epithelium, take a more aggressive form after the modification by tTG2, and trigger a cascade of reactions leading to the intestinal and extra-intestinal symptoms
[14][27][28]. The first reaction initiated by gluten-peptides gets amplified to take a more severe form of an autoimmune disorder upon recognition of tTG2 by the immune system as antigen
[29][30]. The second kind of reaction is gluten-allergy, which involves both the innate and adaptive immune systems
[23][31]. It is a quick reaction against the external allergen within few minutes to hours after ingestion or inhalation and results in a variety of symptoms such as dermatitis, anaphylaxis, etc.,
[32]. The third kind of reaction known as wheat sensitivity involves the innate immune system and is associated with diverse symptoms ranging from fatigue, distress, depression, and migraines to gastrointestinal disorders
[31][33].
The only known treatment for gluten-associated disorders is a life-long wheat exclusion diet
[14][16]. Such a diet is difficult to follow because of the unintended contamination of “gluten-free” products, improper labeling, social constraints, and ubiquity of gluten proteins in raw or cooked food and pharmaceuticals
[34][35][36]. Thus, accidental gluten encounters are likely
[37][38]. Different celiac patients show sensitivity to different gluten proteins
[9][39]. Besides, different individuals show different tolerance levels for gluten intake. In general, celiac patients were shown to tolerate up to 20 mg gluten in a kg of food consumed in a day
[40][41]. Therefore, it is crucial to precisely monitor the gluten content of the food prepared for celiac patients and to maintain gluten-levels below the prescribed limits in their diets
[42].
According to the Codex definition, any food product containing >20 mg/kg gluten cannot be considered or labeled as “gluten-free”
[43]. Because of the gluten contamination, many inherently gluten-free products (derived from corn, rice, millet, oats, etc.,) cannot be consumed by celiac patients
[37][38]. These products, if misbranded as “gluten-free” and used by the celiac patients, will result in recurrence of symptoms
[34]. The gluten contamination can take place at any level from field to the shelf during harvesting, transportation, and/or processing
[44][45][46]. In bakeries where gluten is present ubiquitously, it is almost impossible to decontaminate all equipment, thus unconsciously contaminate the “gluten-free” products
[47]. For instance, in analysis of R5 antibody-based enzyme-linked immunosorbent assay (ELISA) of 22 commonly available, “gluten-free” commodities including grains, seeds, and flours, seven showed mean gluten levels of more than 20 mg/kg
[44]. Antibody-based methods of gluten detection, specifically those relying on R5 and G12 antibodies, are also endorsed by the Prolamin Working Group of the Codex Alimentarius Commission to test gluten contamination in raw and processed food samples (cf.
Table 1)
[48]. There is sufficient evidence to support that even products derived from inherently gluten-free grains cannot be considered safe under the proposed FDA rules for gluten-free labeling
[43][44][49][50][51][52][53][54][55]. As most of these surveys were performed on the raw material, it is very likely, that processed food or convenience products, which have more chances of getting contaminated, will show even higher gluten contamination levels. Besides, most of the existing detection methods employ the sandwich ELISA system, which suffers from the following inherent problems. i) Sandwich ELISA can only be applied to antigens larger than 5-kDa in size and with at least two sterically distant epitopes for their binding and detection by capture and detection antibodies. It thus is unsuitable for the detection of hydrolyzed protein products. ii) The detection limit for most of the assays is 10 mg/kg, with a high error rate close to the lower detection limits. iii) Biased detection of one family of proteins over others, leading to the overestimation of one and underestimation of the other protein family. iv) Protein contaminations in heat-processed food samples (especially glutenins) are difficult to detect in these assays.
Table 1. A list of commonly available gluten detection kits, associated antibodies, target proteins, detection procedures, and extraction systems.
|
Company
|
Neogen Corp.
|
R-Biopharm AG
|
R-Biopharm AG
|
Inmunología y Genética Aplicada SA
|
Romer Labs
|
Tepnel Biosystem
|
Morinaga Inc.
|
|
Product
|
Veratox
|
RIDA-
SCREEN
|
Ridascreen® Gliadin Competitive
|
INgezim Gluten
|
AgraQuant® Gluten G12
|
Gluten assay
|
Wheat protein
|
|
Antibody
|
2 mAb
|
R5 mAb
|
R5 mAb
|
R5 mAb
|
G12 mAb
|
Skerritt mAb
|
Wheat pAb
|
|
ELISA type
|
Sandwich
|
Sandwich
|
Competitive
|
Sandwich
|
Sandwich
|
Sandwich
|
Sandwich
|
|
Time
|
30 min
|
1.5 h
|
40 min
|
60 min
|
60 min
|
30 min
|
2.5 h
|
|
Target
|
gliadin
|
ω, α/β- & γ-gliadins and LMWg
|
ω, α/β- & γ-gliadins and LMWg
|
ω, α/β- & γ-gliadins and LMWg
|
α gliadins
|
ω gliadins and HMWg
|
Wheat proteins
|
|
Antigen
|
|
|
|
|
|
|
|
|
LOD (mg/kg)
|
n/a
|
3
|
1.36
|
3
|
2
|
1
|
0.3
|
|
LOQ (mg/kg)
|
10
|
5
|
5
|
10
|
4
|
10
|
3.12
|
2. Celiac Disease Prevalence
The availability of better tests such as serological analysis for antibodies against tissue transglutaminase (tTG), deamidated gliadin peptide (DGP), and anti-endomysial antibodies (EMA), as well as small bowel biopsy, have improved celiac disease (CD) diagnosis and distinction between CD and non-celiac wheat sensitivity (NCWS)
[26][56][57][58]. Worldwide prevalence of celiac disease has been documented to range between 0.5 to 1.7%
[59][60]. It is noteworthy that the prevalence of CD has increased over the decades, with an incidence rate of 0.6% in the 1990s to 0.8% between 2001 to 2016. Better diagnostics and health awareness might be responsible for this increased CD prevalence. In the northern hemisphere, the incidence in adults diagnosed by biopsy ranges from 0.96% in Canada, 1% in the United States to ~2% in Europe
[61][62]. In the southern hemisphere, the once thought to be the “celiac-free” region of the world has reported having similar CD prevalence to European countries. The latest surveys have established an equal incidence of celiac disease in Asia and Africa
[59][63][64]. Specifically, in India, a celiac disease frequency of 1.04% was reported
[65][66][67]. In some provinces of China, the CD prevalence, similar to the global incidence of two individuals diagnosed every hundred, was reported
[68][69]. In Northern Africa, the CD prevalence of about 0.3 to 5.6% was observed. In Australia and New Zealand, a CD incidence of 0.5% and 1.2 %, respectively, was reported. In South America, specifically Argentina and Brazil, a range of 0.2 to 0.6% was observed. Whereas, in Mexico, a higher rate close to 3% of CD occurrence was reported
[59]. It is possible, over the years, with the advancement in technology and better diagnostics, several patients currently considered to suffer from NCWS will be diagnosed with celiac disease, leading to the changes in our understanding of CD prevalence. The prevalence of NCWS has been documented to range between 0.6 to 13% in the general population. However, because of the lack of diagnostic criteria, biomarkers, and misconceptions on self-diagnosis, the actual prevalence of NCWS cannot be established with a level of confidence today
[70]. These epidemiological studies have suggested that gluten-associated disorders are common around the globe, and reflected toward a need for the “gluten-free” products. However, given the market trend and consumer safety in mind, it is imperative to ascertain that the products labeled as “gluten-free” should have and maintain a gluten-free status.
3. Hidden Gluten or Gluten Contamination
The products containing hidden gluten include sausages, fish fingers, cheese spreads, soups, sauces, mixed seasonings, mincemeats, some medications, and food supplements like vitamin preparations
[34]. Some beverages like real ales, beers, and stout are also contaminated with gluten generally
[36]. The primary causes of contamination are either use of common machinery during the harvest, transportation, and processing or the use of shared storage space
[71]. Mixed, parallel or sequential cultivation of gluten-containing and gluten-free cereals could also lead to contamination of the inherently gluten-free grains
[49]. Contamination is unavoidable if the same milling equipment is used for gluten-free and gluten-containing grains. Interestingly there is no legislature in place regarding the maximum levels of foreign grains in gluten-free cereals. However, in general, 2% of other grains are used as the maximum limit.
The ubiquitous nature of gluten constitutes a global problem. Contamination of food has been reported as early as 1987 in Sweden, United Kingdom, Canada, South Africa, Brazil, the United States, Italy, Australia, and Ireland
[46]. The existence of gluten contamination was reported in industrial and non-industrial products, with or without a “gluten-free” label, with an overall prevalence of 23.2%
[46]. Besides the methodological limitations, such as different detection methods, the experimental design, or non-consensus on the gluten reference material, there is an agreement on the importance of strict regulations to check gluten contamination. Studies conducted over time have shown a decrease in the number of gluten-contaminated samples worldwide
[72][73]. After analysis of a fifteen-year period, more significant decrease was observed in cereals and additives, which were down from 37.7% and 33.0% of gluten contaminated samples to 7% and 2.6%, respectively
[72][74]. In the European Union, 55% of the gluten-free food products were found contaminated in the period between 2003–2005. In contrast, only 19% of the food products were found contaminated in the period between 2013 to 2016, which is concordant with the codex revision implementation followed by Central and Western European countries
[72]. This revision was critical for snacks and yeasts, where gluten level was found unsafe for celiac patients. It forced the yeast bakery product manufacturers to change their practices to ensure gluten control in their products. During recent years, the development of non-invasive methods to detect gluten-related disorders
[75], allowed patients to check the quality of the products and demanding better and reliable food labeling.
4. Available Detection Methods
Over the years, several gluten-detection and quantification methods have been developed and tested using the gluten-containing and/or spiked samples. These procedures can be grossly classified into genomic, proteomic, and immunological methods
[76]. The pros and cons of using these methods are discussed in this section.
The more versatile and commonly accepted assays are immunological assays, in particular ELISA. Owing to the sensitivity and speed of detection, the Codex Committee on Methods of Analysis and Sampling has endorsed these methods
[77]. Several variations of these methods have been developed over the years. Several antibodies (monoclonal and polyclonal) and a variety of commercial kits are available in the market to perform these assays
[76]. The commonly used ELISA systems can be grossly divided into two categories: the sandwich ELISA and the competitive ELISA
[76]. In the sandwich ELISA the antigen is sandwiched between two antibodies, one immobilized to the walls of the microtiter plate (capture antibody) and the other coupled with an enzyme (detection antibody). The sandwich ELISA is only suitable for large antigens because the antigen should have at least two separate epitopes to bind both antibodies. Thus, this ELISA system is not an appropriate choice for partially hydrolyzed gluten samples like in the sourdough products, malt, and beer. The other ELISA system is competitive ELISA, which is suitable for the detection of small-sized antigens with a single epitope. In this system, labeled and unlabeled antigen is applied to immobilized antibody, where they compete for the antibody binding sites. After washing out the unbound antigen, the quantity of the labeled antigen is determined by adding the enzyme-substrate and measuring the intensity of the colored end product, which corresponds with the quantity of the labeled antigen. The major problem associated with both of the ELISA systems is the determination of gluten contamination in heat-processed food samples, which cause conformational changes to the antigen masking or modifying the antibody recognition site(s)
[76]. It has been documented that the α/β- and γ-gliadins lose 49 to 67% of the original reactivity after the heat treatment, while the ω-gliadins remain largely unaffected, i.e., they only lose 7% of reactivity
[78][79]. The commercially available prolamin detection kits are summarized in
Table 1.
The antibody-based detection methods suffer other drawbacks, such as these assays are not fully compatible with the extraction solutions, which lead to the denaturation of proteins
[80]. In recent years, to avoid the limitations associated with antibody-based assays, aptamers were proposed as an alternative. It is generally believed that these molecules can overcome the limitations of using antibodies in the detection, identification, and quantification of specific targets due to their unique properties (cf.
Table 2 and ref.
[81]). The aptamers are single-stranded oligonucleotides that can bind proteins, small-molecules, and living cells with high affinity and specificity
[82]. The single-stranded DNA or RNA oligonucleotide is selected in vitro via a process dubbed as the systematic evolution of ligands by exponential enrichment (SELEX)
[82]. The method relies on the selection of target-specific aptamers through the repetition of the following steps: binding, partition, elution, amplification, and conditioning until the desired aptamer(s) are identified
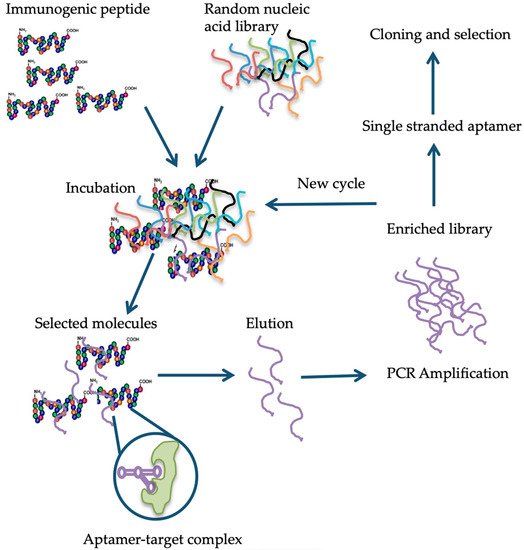
[83]. Briefly, in the case of aptamer designed for gluten detection, specifically the 33-mer immunogenic epitope, a library consisting of 10
14-10
15 single-stranded DNA oligonucleotides with a portion of the random nucleotide sequence is synthesized by a combinatorial chemical synthesis technique, and incubated with the target
[84]. Unconjugated or low-affinity binding molecules are removed, and captured nucleic acid molecules are eluted and amplified by PCR. As a result, double-stranded PCR products are produced, which are later converted to single-stranded aptamers. The whole process is repeated several times until a group of high-affinity binding aptamers is obtained
[85] (
Figure 1). Aptamers are small molecules typically < 100-mers that fold into three-dimensional structures with their self-annealing properties. Target identification is due to their structure and not by their sequence (see
Figure 1). Aptamer-target complexes present dissociation constants (Kd) within the low picomolar (1 × 10
−12 M) to nanomolar (1 × 10
−9 M) range, which reflects toward their high binding affinity. Furthermore, target recognition is highly specific because aptamers can clearly distinguish between closely related protein targets
[86].
Figure 1. Schematic representation of the different steps in the systematic evolution of ligands by exponential enrichment (SELEX) procedure, modified from Banerjee
[87] and Stoltenburg et al.
[83][88].
Table 2. Comparison of aptamers and antibodies based on properties.
|
Properties
|
Aptamers
|
Antibodies
|
Reference
|
|
Affinity
|
Very high target affinity, dissociation constants from micro to picomolar range.
|
Lower target affinity except for some monoclonal antibodies.
|
[89]
|
|
Immunogenic effect
|
Independent of immunogenic effect, due to their in vitro production.
|
Immune response can fail when the target molecule, has a structure similar to an endogenous protein.
|
[90]
|
|
Specificity
|
High binding specificity, e.g., the Anti-theophyllin aptamer displayed 10,000-fold discrimination against caffeine (Theophyllin differs from caffeine by a single methyl group).
|
Depends on target type.
|
[91]
|
|
Production
|
In vitro.
|
In vivo. Use of animals or cell lines.
|
[92]
|
|
Consistency
|
Chemical synthesis, extreme accuracy, and reproducibility. Little or no batch-to-batch variation.
|
May have in vivo variations. Restricted to environmental conditions.
|
[90]
|
|
Properties
|
Can be optimized on demand for increasing binding affinity and specificity.
|
Properties cannot be changed on demand.
|
[84][88]
|
|
Stability
|
Undergo denaturation, but reversible within minutes.
|
Irreversible denaturation. Stable under physiological conditions
|
[90]
|
|
Range of targets
|
Combinatorial library can be produced against any type of target, even toxic targets.
|
Restricted to molecules that produce immunogenic effect.
|
[93]
|
|
Shelf-life
|
Stable to long-term storage at ambient temperature.
|
Limited shelf-life.
|
[92]
|
|
Functionalization
|
Labeling does not affect affinity.
|
Attachment of molecules can cause loss in affinity.
|
[84][88][94]
|