1000/1000
Hot
Most Recent

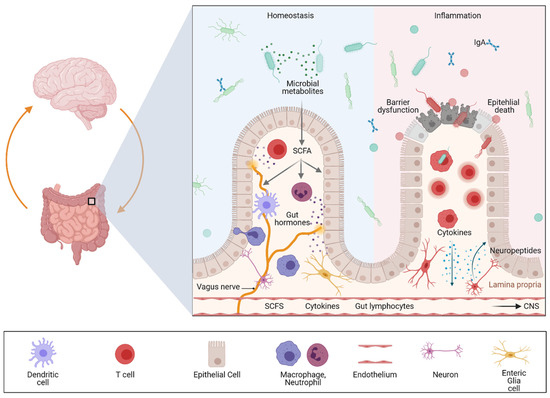
The gut–brain axis is a bidirectional communication system driven by neural, hormonal, metabolic, immunological, and microbial signals. Signaling events from the gut can modulate brain function and recent evidence suggests that the gut–brain axis may play a pivotal role in linking gastrointestinal and neurological diseases. Accordingly, accumulating evidence has suggested a link between inflammatory bowel diseases (IBDs) and neurodegenerative, as well as neuroinflammatory diseases.
The majority of PD cases are of sporadic origin; however, about 10% are familial and the most common monogenic forms of PD are pathogenic variants on the LRRK2 gene that encode leucine-rich repeat kinase 2 (LRRK2) [24], a multidomain protein with a catalytic core that can fulfill kinase and GTPase activity. It also has a scaffold function allowing LRRK2 to interact and recruit several other signaling molecules. The coding variants associated with PD cluster within the enzymatic core of LRRK2 and are thought to disrupt the enzymatic functions of this protein. Accordingly, preclinical studies have indicated that targeting the activity or expression of LRRK2 is neuroprotective.
Recent genome-wide association studies (GWAS) have shown that the association between the LRRK2 locus and IBD includes several LRRK2 genetic variants [25]. Although the interaction of alterations in LRRK2 function and CD mechanisms is still unknown, increasing evidence supports the fact that LRRK2 plays a role in mediating autophagy in Paneth cells, which would explain its strong association with CD because defects in Paneth cell autophagy have been described as hallmarks of this IBD prototype [26]. In line with this notion, preclinical studies using mice with a LRRK2 deficiency showed a specific impairment in the expression of antimicrobial peptides by Paneth cells [27].
The genetic risk factors associated with MS and IBD are not well described in the literature. Genetic studies of MS cohorts suggest that this autoimmune disease is provoked following exposure to environmental factors, which might be responsible for loss of tolerance and peripheral activation of myelin-specific T cells. GWAS have supported the complexity of MS pathology and uncovered immune-related gene variants linking MS to other autoimmune diseases such as IBD [28]. Further systematic studies are needed to better delineate the genetics between IBD and MS.
Beside the epidemiological and genetic evidence, several preclinical studies support the importance of gut–brain communication in intestinal inflammation. In the dextran sulfate sodium (DSS)-induced colitis, a rodent model of IBD, it was demonstrated that, parallel to local inflammatory responses in the gut mucosa, increased expression of IL6 and iNOS (Nitric oxide synthase, inducible; NOS2) was found in the cerebral cortex. The authors further described microglial activation by increased immunoreactivity for the pan-myeloid cell marker ionized calcium-binding adapter molecule 1 (Iba1) and elevated cytokine levels [29]. Another study analyzed the impact of intestinal inflammation by DSS administration in a model of dopaminergic neurodegeneration by LPS injection in the substantia nigra [30]. The authors demonstrated that inflammatory responses in the gut reinforced the inflammatory and deleterious effects of LPS induced neuroinflammation as indicated by increased levels of TNF-α, GFAP, and IL-6 in serum and the substantia nigra of the animals.
Interestingly, there are several recent animal studies suggesting that α-synuclein, a key protein involved in PD pathology, accumulates not only in the brain but also in the gut. Surprisingly, this could not only be observed in α-synuclein transgenic mice but also in mice subjected to DSS (experimental colitis drives enteric α-synuclein accumulation and Parkinson-like brain pathology). In line with previous examinations, this study observed that experimentally induced colitis in transgenic mice exacerbated α-synuclein pathologies in the CNS. While highly interesting, the effect of α-synuclein aggregates on ENS homeostasis has not been studied. So far, only one study demonstrated that increased α-synuclein expression following colitis was associated with phosphorylation in the myenteric plexus of common marmosets [31].
In summary, available studies suggest that inflammation is associated with peripheral alterations affecting CNS homeostasis through factors accumulating in the gut or systemically. In line with this hypothesis, another recent study demonstrated activation of microglial cells and reduction in occludin and claudin-5 expression in the brain suggesting an impaired BBB following experimental colitis [32]. These data further suggest that DSS-induced colitis increases systemic inflammation which then results in cortical inflammation via up-regulation of serum cytokines. Interestingly, the same group further observed that a decrease in dopaminergic function was associated with an increase in gastrointestinal inflammation, suggesting a bidirectional gut–brain interaction. Accordingly, mice studies showed that Experimental Autoimmune Encephalomyelitis (EAE), a model for MS in rodents, is accompanied by loss of mucosal immune homeostasis [33].
The GI tract is the only internal organ that has its own independent nervous system, the enteric nervous system (ENS) [34][35]. This digestive system can be innervated by intrinsic enteric neurons and by extrinsic efferent and afferent nerves.
It has been shown that neuropeptide-containing (peptidergic) neurons within the colonic wall are key players in neurogenic inflammation as they release neuropeptides into the adjacent tissue. These peptides can induce vasodilation, plasma extravasation and leukocyte migration. Moreover, these neuropeptides have been shown to not only regulate intestinal homeostasis but also inflammation [36]. Accordingly, experimental studies could demonstrate that peptidergic neurons release neuropeptides that orchestrate colonic inflammation in a complex way. Calcitonin gene-related peptide (CGRP) and substance P (SP) seem to be the link between neuronal activation and the consecutive mucosal immune response. In murine colitis models, mice deficient in neutral endopeptidase (an enzyme responsible for the extracellular degradation of SP) displayed and aggravated colitis, while mice deficient in substance P showed a strong attenuation of colitis severity [15][37][38]. In sharp contrast, CGRP-deficient mice showed increased susceptibility to experimental colitis. Neuropeptide release is controlled by transient receptor potential (TRP) channels; therefore, neuropeptides released locally in the gut may function as mediators at the interface between the nervous system, the mucosal immune system and other cell compartments such as the epithelium or endothelium. Interestingly, recent single cell analyses revealed a significant expression of risk genes for diseases that feature intestinal and CNS involvement in the ENS, suggesting that it is involved in gut–brain disease communication [16].
While the importance of the gut microbiome was described some time ago for IBD and a variety of other immune and metabolically driven diseases, the essential role of the gut microbiota in CNS inflammation was discovered only a few years ago [39].
Several clinical studies highlighted a reduced diversity and altered composition of the gut microbiota (dysbiosis) not only in mouse models of neuroinflammation and neurodegeneration, but also as a common feature of patients with PD [40][41][42][43][44] and MS [45][46][47][48][49][50][51][52]. While dysbiosis has been shown in many clinical and preclinical studies, a disease-relevant microbiota for neuroinflammation or neurodegeneration is debatable. In addition, it still remains unclear whether dysbiosis can modulate inflammatory processes in the CNS or if it is merely the consequence of neuroinflammation/neurodegeneration. In support of a rather causative function of gut microbes, germ-free mice were resistant to spontaneous EAE, a striking notion that was explained by a lack of local activation of T cells in the gut and the subsequent deficient triggering of pathogenic antibody production by activated B cells. A translational study could demonstrate that transplanting faecal microbiota from PD patients exacerbated motor dysfunction in an α-synuclein transgenic mouse model [53].
In summary, there is growing evidence suggesting that the gut is strongly involved in various neurological diseases via direct and indirect mechanisms. The key components are intestinal microbes and their products (e.g., metabolites) and immune education in the mucosal immune system, including immune cells releasing proinflammatory cytokines. Key to the regulation of these processes is the intestinal epithelium, which is capable of translating microbial and inflammatory signals to the immune system and secreting peptides as well as hormones, which are involved in the metabolic processing of dietary nutrients. Although this network is of strong clinical relevance for both intestinal and neurological diseases, we are just beginning to understand the underlying molecular mechanism and how organ crosstalk is regulated during health and disease. Accordingly, we need novel model systems to better understand microbiota–gut–brain communication on a cellular level. In the last chapter we therefore focus on novel human-specific preclinical model systems that will help to uncover disease mechanisms, which might allow us to better understand and modulate the function of this complex system.