1000/1000
Hot
Most Recent

Enteroviruses (EVs) are positive-sense RNA viruses, with over 50,000 nucleotide sequences publicly available as of October 9th, 2020. While most human infections are typically associated with mild respiratory symptoms, several different EV types have also been associated with severe human disease, especially acute flaccid paralysis (AFP), particularly with endemic members of the EV-B species and two pandemic types—EV-A71 and EV-D68—that appear to be responsible for recent widespread outbreaks. Phylogenetic analysis of the EV genus shows the evolutionary relatedness of different EV types.
Enteroviruses (EVs) are members of the Picornaviridae family of small (30 nm diameter virions) non-enveloped viruses. The enterovirus genome is a single-stranded positive-sense RNA molecule of approximately 7500 nucleotides, containing a 5′ untranslated region (UTR) of approximately 750 nucleotides and a short 3′UTR of approximately 70–100 nucleotides[1]. Enteroviruses include important human pathogens, including polioviruses, rhinoviruses, echoviruses, and coxsackieviruses. Established in 1988, the World Health Organization (WHO) Global Polio Eradication Initiative (GPEI) contributed to an initial wealth of information on various enteroviruses[2]. In addition to polio surveillance, non-polio enteroviruses are passively surveilled across several governmental and intergovernmental agencies, primarily across the Asia–Pacific region, Europe, and the United States. Enteroviruses typically cause mild respiratory and/or gastrointestinal diseases. However, they can also be associated with severe disease, even in otherwise healthy individuals[1]. Here we review the evolution and phylogenetic relationship between the non-polio and non-rhino enterovirus clades and explore their disease associations.
Enteroviruses are classified into 15 species by the International Committee on Taxonomy of Viruses (https://talk.ictvonline.org)—enterovirus (EV) A through L and rhinovirus (RV) A through C. RV-A, B, and C and EV-A, B, C, and D are the only enterovirus species that infect humans. Enterovirus subspecies were originally classified serologically based on cross-neutralization testing and were referred to as “serotypes”. More recently, due to the widespread use of genome sequencing for virus characterization, subspecies classification is now based on the sequence of the major hypervariable capsid protein VP1 and they are referred to as “types”. To date, 326 enterovirus types have been identified across the 15 different species. Among the different species that infect humans, these include 168 different rhinovirus types, 3 poliovirus types, and 103 other enterovirus types[3]. Enterovirus types are defined based on >75% nucleotide and >85% amino acid sequence identity to a prototype strain[4], although some isolates within a type have drifted slightly beyond these thresholds[5]. New enterovirus types are designated using the prefix EV or RV, followed by the species letter and a type number designation. For historical reasons, some enterovirus types have been referred to differently. Echoviruses are all part of the enterovirus B species; however, they are abbreviated with the prefix “E” (e.g., E30) even though they are not members of the enterovirus E species. Specific coxsackievirus (CV) types are designated with the CV prefix and can belong to either EV-A, EV-B, or EV-C species depending on their VP1 sequence characteristics.
Like other positive-sense RNA viruses, enteroviruses encode a polymerase that lacks a proof-reading mechanism[6], and they are one of the most rapidly evolving virus groups[7], with a relatively high substitution rate estimated to be 5–12 × 10−3 substitutions per nucleotide per year (s/n/y)[5][8]. We performed a phylogenetic analysis of VP1 nucleotide sequences which illustrates the species and type relationships of human enterovirus isolates (Figure 1). The Virus Pathogen Resource (ViPR; https://www.viprbrc.org)[9] was used to collect enterovirus sequence data for this analysis. ViPR is part of the Bacterial and Viral Bioinformatics Resource Center (BV-BRC), a US National Institute of Allergy and Infectious Diseases (NIAID)-sponsored bioinformatics repository of data and analysis tools for major human viral pathogens, including enteroviruses[9].
The four human EV species (A–D) and the three RV species (A–C) are well segregated with 100% bootstrap support values. Isolates of the EV-D species are more similar to EV-A isolates than isolates of the other enterovirus species, whereas EV-C and EV-B isolates are more similar to each than to isolates of the other species, in agreement with previous reports[10]. Type relationships are also reflected in the branching structures with independent isolates of a given EV type closely connected with short branch lengths. For example, all six CV-B3 isolates are located within the same phylogenetic branch even though they come from dispersed geographic regions (India, China, Australia, and France). The tree also highlights the challenge with the historical nomenclature in that echovirus and coxsackievirus types are interspersed in the phylogenetic tree. For example, CV-A9 is quite similar in sequence to E19, while quite different from CV-A10. Thus, the current nomenclature that distinguishes enteroviruses, echoviruses, and coxsackieviruses does not reliably reflect the evolutionary relatedness illustrated in the phylogenetic analysis of VP1 and other genomic regions.
Enterovirus A types are the most common cause of HFMD epidemics. HFMD is characterized by painful sores that can develop in the mouth (herpangina), as well as skin rashes on the palms of the hands and soles of the feet. Initial symptoms include fever, sore throat, and malaise. HFMD epidemics have been increasingly reported across the Asia–Pacific region, particularly in China[11][12], where approximately 1.3 million cases of HFMD were reported in 2009, and that numbers continued to rise until 2014, when over 2.7 million cases of HFMD were reported[13]. Within the EV-A subtree, CV-A16 and EV-A71 are closely related to each other, forming a monophyletic group with a support value of 99%. The genetic similarity between these two types is likely responsible for the fact that both cause HFMD. On the other hand, the clear segregation of CV-A16 and EV-A71 in the tree also correlates with their distinct clinical characteristics. CV-A16 infections are usually mild, while cases of HFMD with severe neurological complications are usually associated with EV-A71 infections. Surveillance studies on HFMD outbreaks found that CV-A16 and EV-A71 often co-circulate[14][15], which could be the result of a common evolutionary origin. Co-circulation of these viruses further provides an ecological niche for intertypic recombination, resulting in intertwined evolutionary trajectories and emergence of new enterovirus variants.
Another monophyletic group within the EV-A subtree includes CV-A2, CV-A4, CV-A5, CV-A6, and CV-A10. Compared with other groups of similar sizes, this group has a larger number of types. This is likely the outcome of finer granularity of virus classification, rather than more genetic diversification. All virus types within this subtree are notably distinct from each other, as shown by the type-level clustering with 100% support values. Among the various types, CV-A2, CV-A6, and CV-A10 have been increasingly associated with HFMD, herpangina, and onychomadesis[16][17][18][19][20]. Similar to the CV-A16/EV-A71 group, CV-A6 and CV-A10 often co-circulate. Furthermore, it has been reported that CV-A6, CV-A10, CV-A16, and EV-A71 co-circulate together[21], which helps to shape the interacting evolution of these viruses.
Enterovirus B is a broad and diverse species that includes 59 different types. Acute flaccid paralysis (AFP) is one of the most serious conditions attributed to enterovirus B. In a systematic review of the literature, enterovirus B types have been implicated in 72% of all enterovirus-associated AFP cases[22]. Echovirus 30 is a common enterovirus B type and is the principle cause of viral meningitis and encephalitis in temperate climates, particularly in Europe, the Americas, and the Asia–Pacific. Numerous other EV-B types have also been associated with severe diseases, including encephalitis and meningitis[10]. Myocarditis has been associated with a number of EV-B types, most significantly the CV-Bs[23][24]. In the EV-B subtree, each type forms its own distinct cluster. Compared with EV-A and EV-C, different EV-B types are more similar to each other. Consequently, not all intertypic clusters in this subtree have strong support values. One of the major pathogens in EV-B is E30, which has caused large outbreaks in Asia, Europe, and Americas. The four E30 strains in the subtree form a tight cluster (bootstrap = 100%) with diverse year coverage. Studies have shown the independent evolution of structural and non-structural gene regions in EV-B[25]. The VP1 region experiences progressive drift as lineages emerge, dominate, and then are replaced by new lineages over periods of 3–5 years. The temporal diversity in the E30 cluster is congruent with the lineage replacement model. It should be noted that the percentage of complete genomes/all genomes of EV-B is significantly lower (1.8%) than that of EV-A (5.2%), EV-C (5.5%) or EV-D (12.1%). Due to the small number of complete genomes available for E30, the E30 sequences used in this study may not adequately reflect the overall type diversity, even though they still cover the Asia–Pacific and America regions, and different years. With this limitation in mind, the representative E30 sequences selected by clustering still show a temporal rather than geographic relationship, which can be explained by the time-related turnover model[25].
Non-polio enterovirus C types are relatively uncommon. CV-A24 has been responsible for outbreaks of conjunctivitis—an inflammation of the outer membrane of the eyelid[26][27][28]. The EV-C subtree has three major monophyletic groups. The poliovirus group is farther away from the rest of the EV-C viruses, which is consistent with the fact that poliovirus has higher incidence of paralysis than other types. CV-A21, CV-24, EV-C96, and EV-C99 belong to the same group. Even though they are genetically close, they exhibit a wide spectrum of clinical manifestations, with CV-A24 causing conjunctivitis, and C96/C99 associated with AFP and gastroenteritis.
Enterovirus D infections were not frequently reported prior to 2012. However, that changed with the EV-D68 biennial outbreaks starting in 2014. EV-D68 was first detected as a respiratory virus in children with pneumonia and bronchiolitis in 1962[29]. Since then, a number of distinct clades have evolved and are currently co-circulating worldwide[30]. In the summer and fall of 2014, North America experienced a widespread outbreak of severe respiratory illness associated with EV-D68, with smaller clusters of infections also reported in Europe and Asia, resulting in the public health community classifying EV-D68 as a re-emerging pathogen of public health concern[31] . EV-D68 cases peaked again in 2016 and 2018 following a biennial pattern typical of some enterovirus types, particularly in the US[32][33][34][35][36][37]. In the 2014 outbreak, reports of acute flaccid myelitis (AFM) (AFP with radiographic evidence of spinal cord inflammation) in some children with detectable EV-D68 raised concerns that genetic changes in EV-D68 could be contributing to increased disease severity and neurological symptoms[38]. A report on the Bradford Hill criteria for establishing a causal relationship in human infectious diseases showed that EV-D68 fulfills seven of the nine criteria[39]. In particular, the co-clustering of EV-D68 infections and AFM in the summer/fall of the 2014, 2016, and 2018 outbreaks[36][40][41] suggests a causal relationship. Due to the lack of whole-genome sequences for EV-D70 and EV-D94, the EV-D species is currently only represented by a single type—EV-D68—in this phylogenetic analysis.
In conclusion, although poliovirus and rhinovirus may be more widely recognized as human infectious disease agents, the non-polio and non-rhino enteroviruses are important global endemic and epidemic pathogens. Their wide genetic diversity makes it challenging to develop sensitive and specific diagnostic reagents or cross-protective preventative vaccines. The fact that enteroviruses can often cause severe and a diverse array of diseases in humans motivates their further study by the research community.
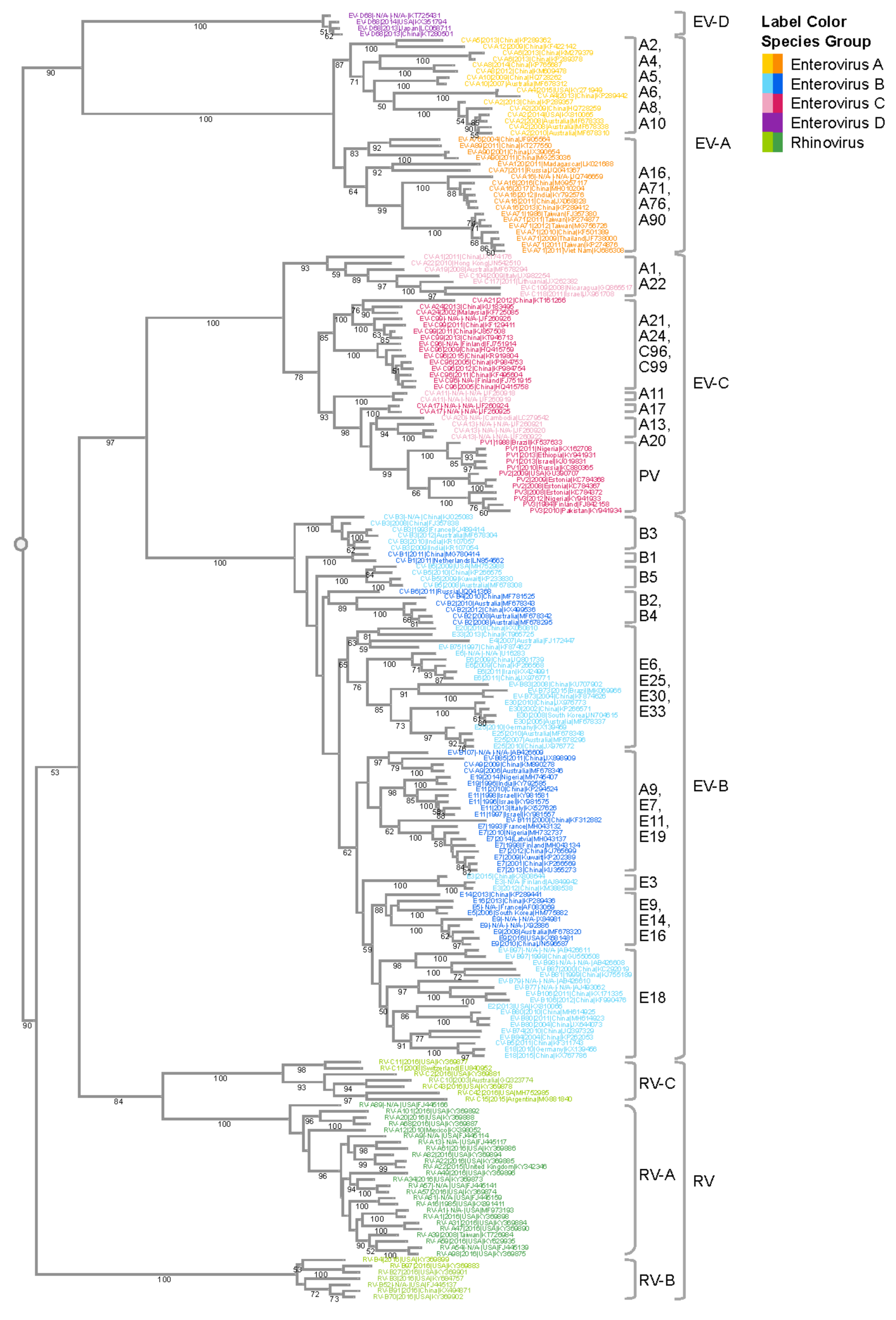
Figure 1. Phylogenetic analysis of the enterovirus genus. VP1 gene sequences from complete human enterovirus genomes were retrieved from the ViPR database (https://www.viprbrc.org/). Sequences representative of the genomic diversity were selected using CD-HIT[42], with a threshold of 0.90, and subsequently aligned using MAFFT E-INS-i. The phylogenetic tree was generated using RaXML with 100 bootstrap replicates, and then visualized in Archaeopteryx.js on ViPR with leaf nodes color coded by phylogenetic relatedness. Bootstrap support values of 50 or larger are shown in the tree. Major monophyletic groups are labeled with the major types in the group. Major types are defined as those with 50 or more genomes in the case of EV-A and EV-C, and 200 or more genomes in the case of EV-B.