1000/1000
Hot
Most Recent

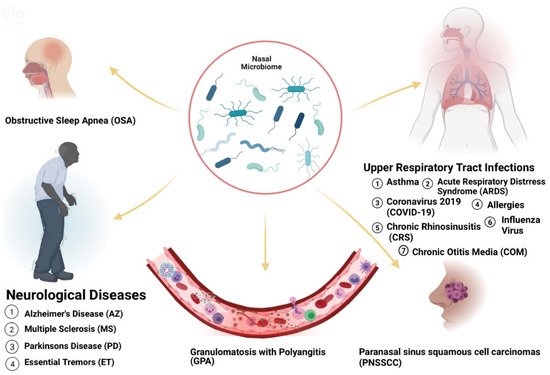
Acute and chronic upper respiratory illnesses such as asthma, and allergic rhinitis (AR) have been linked to the presence of microorganisms in the nose. Microorganisms can exist in symbiotic or commensal relationships with the human body. However, in certain cases, opportunistic pathogens can take over, leading to altered states (dysbiosis) and causing disease. Thus, the microflora present in a host can be useful to reflect health status. The human body contains 10 trillion to 100 trillion microorganisms. Of these populations, certain pathogens have been identified to promote or undermine wellbeing. Therefore, knowledge of the microbiome is potentially helpful as a diagnostic tool for many diseases. Variations have been recognized in the types of microbes that inhabit various populations based on geography, diet, and lifestyle choices and various microbiota have been shown to modulate immune responses in allergic disease. Interestingly, the diseases affected by these changes are prevalent in certain racial or ethnic populations. These prevalent microbiome variations in these groups suggest that the presence of these microorganisms may be significantly associated with health disparities.
| Disease | Greater Abundance Linked to Disease | Affected Group | Known Risk Factors |
|---|---|---|---|
| Chronic otitis media with effusion (COME) | Corynebacterium, Streptococcus, Moraxella [16] | Caucasian Americans (CAs) [17] | allergy or atopy, upper respiratory tract infection, snoring, acute otitis media, passive smoke exposure and low social status [17][18] |
| Pediatric Asthma | Corynebacterium [19] | Puerto-Ricans (PRs), African Americans (AAs) [20] | parental asthma, prenatal environmental tobacco smoke, having cats and prematurity [21][22] |
| Chronic Rhinosinusitis (CRS) | Corynebacterium, Burkholderia [23][24] | Latino Americans (LAs) [24] | Asthma, genetics, GERD, rheumatoid arthritis, migraine, cigarette smoking [25] |
| COVID-19 | Salmonella, Scardovia, Serratia and Pectobacteriaceae [26][27] | AAs and Asian Americans (AS) [28] | Hypertension, coronary artery disease, history of stroke, diabetes, obesity, severe obesity, chronic kidney disease, asthma, and chronic obstructive pulmonary disease [29] |
| Granulomatosis with Polyangiitis (GPA) | Staphylococcus aureus [30] | Not Distinguishable [31] | Animal (horses) exposure, history of bronchiectasis, autoimmune disease, chronic renal impairment, Pulmonary fibrosis [32][33] |
| Acute Otitis Media (AOM) | Haemophilus, Moraxella, and Neisseria [16] | CAs [34] | Out-of-home daycare, multiple children living in the home, and mother’s multiparity [34] |
| Influenza B virus (IBV) | Streptococcus species and Prevotella salivae [35] | Diabetes, chronic respiratory disease [36] | |
| Influenza A virus (IAV) | Staphylococcus aureus, Staphylococcu pneumoniae, Klebsiella and Aerococcus [37] | ||
| Acute Respiratory Distress Syndrome (ARDS) | CAs [38] | abuse of alcohol and tobacco, malnutrition and obesity [39] | |
| Obstructive Sleep Apnea (OSA) | Streptococcus, Prevotella and Veillonella [40] | Obesity, rhinitis, adenoid hypertrophy [41] | |
| Parkinson’s Disease (PD) | Actinobacter [42][43] | OSA, Head injury, family history of trauma and depression, family history of PD [41][44] | |
| Alzheimer’s Disease (AZ) | OSA, diet, the immune system, mitochondrial function, metal exposure, and infection [41][45] | ||
| Essential Tremors (ET) | AAs, LAs [46] | Exposure to neurotoxic compounds such as β-carboline alkaloids and ethanol. Exposure to pesticide and lead. Tobacco exposure [47][48] | |
| Atopic Dermatitis (AD) | AAs [49] | Viral and bacterial skin infections, neuropsychiatric diseases, family history, smoking, allergy, maternal asthma, and dogs [50] | |
| Psoriasis | CAs, Eastern African [51][52] | Stress, diabetes mellitus, obesity, smoking, air pollution arthritis, inflammatory bowel disease, alcohol, drugs, cardiovascular disease, infection, sun exposure, hypertension [53][54] |