1000/1000
Hot
Most Recent

microRNAs are small noncoding RNAs that regulate gene expression at the posttranscriptional level. Let-7d is a microRNA of the conserved let-7 family that is dysregulated in female malignancies including breast cancer, ovarian cancer, endometrial cancer, and cervical cancer. Moreover, a dysregulation is observed in endometriosis and pregnancy-associated diseases such as preeclampsia and fetal growth restriction.
MicroRNAs are a group of endogenous small single-stranded non-coding RNA molecules of 19–24 nucleotides that regulate gene expression at the posttranscriptional level [1][2][3][4]. Most microRNA encoding genes are inserted in intronic regions of protein-encoding genes. They are either transcribed under the control of their own promoters or they share a promoter with mRNA. In the genome, microRNAs can be organised as microRNA clusters [1]. After transcription by RNA polymerase II in the nucleus, primary micro-RNA (pri-microRNA), comprising about 500–3000 nucleotides, is processed into stem-loop precursor microRNA (pre-microRNA) by RNA III endonuclease Drosha and the dsRNA binding protein DGCR8/Pasha (a complex known as microprocessor) [4]. After translocation into the cytoplasm via the transport system exportin-5 in a Ran-GTP-dependent process, pre-microRNA is converted into a ~20-bp microRNA duplex by RNAse III-enzyme Dicer characterized by the presence of a 5′ phosphate and a two nucleotide 3′ overhang on each end [4][5]. Of the microRNA double strand, the mature microRNA enters the guide-strand channel of an Argonaute protein thus forming a silencing complex, whereas the complementary microRNA* strand is degraded. While binding to 3′-untranslated regions (UTR) of complement target mRNAs as part of the RNA-induced silencing complex (RISC) microRNAs promote either their degradation or translational repression, depending on the degree of complementarity between the so-called seed-sequence of the mature microRNA and the corresponding complementary target region in the cognate mRNA [2][3][6]. MicroRNAs are expressed in tissue and cell-type-specific expression profiles and they are implicated in physiologic and pathologic processes, including the cell cycle, apoptosis, proliferation, differentiation, metabolic pathways and cell response to various types of stress [7][8][9][10].
The human lethal-7 (let-7) family has an important role in the regulation of cell proliferation and carcinogenesis. It includes 13 members in 9 loci on 7 chromosomes, and represents an evolutionarily conserved microRNA family [11].
The mature let-7 family members are the most common among all microRNAs [4]. Let-7d is one of the members of this family, and is located within the let-7a-1/let-7f-1/let-7d cluster on chromosome 9, which is transcribed as a single polycistronic transcript [11]. Let-7 expression is controlled by different transcriptional and post-transcriptional processes, including transcriptional regulation by OCT4, MYC and mutant p53 [11][12], and posttranscriptional regulation by the RNA binding protein LIN28 [13], and the RNA editing enzyme ADAR [14] ( Figure 1 ).
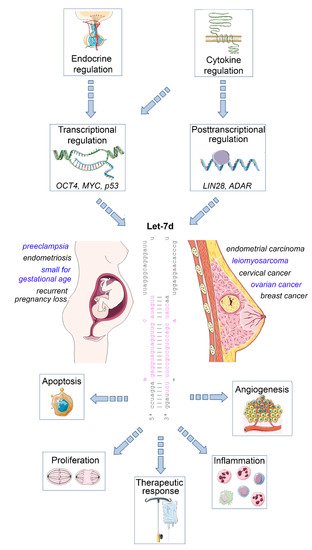
An important let-7 target is the RAS family, regulating K-RAS mRNA and therefore the cell proliferation is reduced upon let-7d upregulation. Other targets are MYC, IMP-1 and High Mobility Group A2 (HMGA2) [16]. In line with this tumor-suppressive role, let-7 family members are de-regulated in several human cancers. The expression of let-7d is low in cancer as pancreatic, prostate, primary pigmented nodular adrenal dysplasia, head and neck, bladder, and kidney cancer [17][18][19]. In a therapeutic context, after irradiation, let-7 family members can be up- and downregulated, which depends on dosage, time after the irradiation, source of oxidative stress and genetic background of the cell [20][21]. Apart from acting on tumor cells, let-7d may also regulate the inflammatory tumor microenvironment, as an immunoregulatory role has been demonstrated for this microRNA. For example, transcriptional analysis and microRNA inhibitor studies have demonstrated that exosomal delivery of Let-7d from Treg cells to Th1 cells was able to suppress systemic inflammatory disease in a mouse model [22]. Moreover, studies in knockout mice revealed that the let-7 cluster (including let-7d) suppresses B cell activation via a metabolic mechanism that resulted in restriction of necessary nutrients, thereby affecting B-cell dependent antibody production [23]. Finally, let-7d targets the epigenetic regulator Tet2, which demethylates deoxycytosine residues in DNA, resulting in enhanced expression of IL-1ß and IL-6 in macrophages in murine models of inflammation [24]. Overall, these studies emphasize the relevance of let-7d as an important suppressor of tumor progression and regulator of inflammatory processes. In the following sections, we will highlight the role of dysregulated let-7d expression in female malignancies and diseases of the female reproductive tract ( Figure 1 ).
Impact of let-7d on female malignancies and diseases of the female reproductive tract. Let-7d expression is dysregulated in a variety of gynaecological and obstetric disorders. Its expression is regulated in a hormone- and cytokine-dependent manner involving both transcriptional and post-transcriptional mechanisms. Let-7d targets key mRNAs involved in the regulation of cell proliferation, apoptosis, angiogenesis and immune cell function, thereby modulating disease progression and therapeutic response. See text for details. The Let-7d sequence was retrieved from miRbase [15]. This figure was designed using elements of the free web resource Smart Servier Medical Art ( https://smart.servier.com/ (accessed on 1 June 2021)).
Ovarian cancer is the seventh most frequent cancer and the eighth main cause of cancer-associated mortality [25]. The prognosis is poor: In the majority of cases the disease is in an advanced state at the moment of the diagnosis and the overall 5-year survival rate is only about 50%. Ovarian cancer has different histologic types; the most frequent is epithelial ovarian cancer (with the subtypes: serous, endometroid, mucinous and clear cell). Most of the patients have high-grade serous ovarian cancer, distinguished by an aggressive development and poor prognosis [26]. Surgery is important not only for the diagnosis and staging of ovarian cancer, but also in the treatment management of patients with advanced disease [27]. Platinum-based chemotherapy (including cisplatin and carboplatin) is the first-line agent in the treatment of ovarian cancer. Metastases are the main reason for the high mortality in these patients [28]. Studies in model organisms have revealed a gradual increase in ovarian and pituitary let-7 expression, including let-7d, during ovarian development, suggesting a potential role of this microRNA in ovarian function [29].
Similar to other tumor entities, aberrant expression of let-7 family members in patients with ovarian cancer compared with that in healthy controls was identified in several studies and so the differentially expressed microRNA may have potential as a diagnostic marker [30][31][32]. However, let-7 dysregulation showed a heterogenous pattern, depending on the subtype and family member involved. For example, in liquid biopsies, let-7b is upregulated, whereas let-7f and let-7i are downregulated in ovarian cancer [33]. Among all let-7 family members, also let-7d was found to be dysregulated in ovarian cancer [34], and it has been shown that the cytokine PDGF-AA is capable of repressing let-7d expression in ovarian cancer cells [35]. High throughput sequencing revealed that the level of plasma exosomal let-7d-5p in patients with ovarian cancer was significantly higher compared to the controls [30]. Similarly, let-7d-3p was upregulated in ovarian cancer tissue relative to normal ovarian tissues in a study assessing its role in the response to neoadjuvant chemotherapy [31]. Moreover, serum levels of let-7d-3p were found to be able to discriminate epithelial ovarian cancer patients from healthy controls [32]. In the same study, HMGA2 and Kirsten Rat Sarcoma Viral Oncogene Homolog (KRAS) were identified as predicted targets of let-7d by bioinformatics analysis, and their expression levels also had a diagnostic value in the studied patient collective [32]. HMGA2 regulates gene expression by binding to AT-rich regions of DNA and thereby promotes tumor progression via different mechanisms. These include the promotion of a cancer stem cell phenotype, improved DNA repair mechanisms linked to therapeutic resistance, and regulation of multiple signaling processes that drive prometastatic EMT [36][37][38]. KRAS also has an ongogenic function in ovarian cancer, as constitutive activation of this small GTPase by gene mutations activates uncontrolled ovarian cancer cell proliferation via activation of the mitogen axctivated protein kinase (MAPK)/ extracellular signal regulated kinase (ERK) pathway, a signaling pathway that is also regulated by HMGA2 [36][39]. Therefore, downregulation of HMGA2 and KRAS by let-7d could exert an anti-oncogenic effect in ovarian cancer. However, a study comparing epithelial ovarian cancer cell lines and immortalized ovarian surface epithelium cell lines reported a significant decrease of let-7d in the cancer cells, suggesting potential cell-type- or context-dependent variability regarding let-7d expression in this tumor entity [40]. Aberrant let-7d levels are not only seen in ovarian cells, but also in cells of the tumor microenvironment, as let-7d was significantly downregulated in highly inflamed lymphatic vessels infiltrating ovarian tumors compared to less inflamed vessels [41]. Notably, this study revealed that high inflammation correlated with the relapse of ovarian cancer.
A variety of mechanistic studies supports the concept of a specific functional involvement of let-7d in ovarian cancer pathogenesis. Analysis of a panel of patient-derived high-grade serous ovarian cancer cells revealed an inverse association between microRNA let-7 expression and the cancer stem cell phenotype, with low let-7 levels being associated with an epithelial phenotype [42]. This finding may be associated with a regulatory impact of the EMT-marker snail on let-7 [43]. One target of let-7d is c-Myc, an essential oncogenic transcription factor that is involved in tumor pathogenesis and frequently dysregulated in cancer [44]. c-Myc expression is upregulated in ovarian cancer, driving tumor progression by regulating the expression of genes that control tumor cell proliferation, cell-cycle progression, angiogenesis, apoptosis, as well as cell adhesion and motility as a prerequisite for metastatic spread [45]. Let-7d suppresses c-Myc and increases ovarian cancer cell sensitivity to 7-difluoromethoxyl-5,4′-di-n-octylgenistein (DFOG) by inhibiting the PI3K/AKT pathway [46]. DFOG is a synthetic genistein analogue that is under preclinical evaluation. It exerts an anti-ovarian cancer effect by repressing tumor stemness via suppression of PI3K/AKT signaling in vitro and in vivo [46]. In another study, it was demonstrated that an additional target of let-7d is high mobility group A1 (HMGA1) [47]. HMGA1 controls the p53 signaling pathway that is important in the regulation of cell apoptosis and drug resistance at the transcriptional level [48][49]. A putative regulatory axis of let-7d, HMGA2 and KRAS may be associated with tumorigenesis, invasion, and metastasis in epithelial ovarian cancer [32].
Several studies point to a link between let-7d expression and drug response in ovarian cancer [31][47][49]. Indeed, serum levels of let-7a have been proposed as a biomarker for treatment decisions regarding the type of chemotherapy (platinum vs. platinum plus paclitaxel) [33][50]. Considering let-7d, let-7d-5p inhibits ovarian cancer cell motility, cell cycle progression and promotes cell death and cisplatin chemosensitivity by repressing HMGA1 via the p53 pathway [48]. Moreover, an increased expression of let-7d-3p was associated with a better response to carboplatin/paclitaxel treatment in ovarian cancer patients [31]. Functional evaluation of let-7d-3p in SKOV3 ovarian cancer cells in this study revealed that its inhibition impaired cell proliferation and activated apoptosis, but did not affect cell motility and invasiveness. While these studies point at an important role for let-7d dysregulation in ovarian cancer pathogenesis, some context-dependent effects of let-7d function need to be taken into account to evaluate the side effects of potential let-7d-centered therapeutic approaches.
The process of reproductive aging has strong implications for female fertility and reproductive outcome, not only from a clinical, but also from a molecular to a morpho-functional point of view [51]. Indeed, an advanced reproductive-aged female phenotype is associated with altered expression of specific microRNAs, regulating gene expression, chromatin remodelling and early embryo development, as summarized in a recent review [52]. While let-7d or other let-7 family members were not specifically identified in these studies, future studies in this context may be worthwhile, as research on animal models has identified a clear link between let-7 and aging in general. In the worm C. elegans , let-7 regulates developmental timing, and low levels of the RNA-binding protein LIN-28 enhance longevity, and reduce germline progenitor cell numbers by regulating let-7, which in turn targets the signaling kinases Akt-1/2 and the downstream transcription factor DAF-16 [53][54]. Notably, in the fruit fly Drosophila , let-7 regulates ageing of the testis stem-cell niche, indicating its relevance for male reproduction [55]. Finally, studies in knockout mice deficient in the long noncoding RNA H19 have suggested a possible regulatory impact of let-7 on anti-muellerian hormone (AMH): H19 acts as a “sponge” inhibiting let-7 function, and let-7 was demonstrated to target AMH expression [56]. In the mouse model, absence of H19 resulted in subfertility, accelerated follicular recruitment and decreased ovarian AMH levels. As AMH is an important marker of the ovarian reserve and the age-related decline of the ovarian pool, a more detailed investigation of the role of let-7 family members in female reproduction could be a promising approach [57][58].