rDNA-mediated translocations occur at about the same frequency in the normal T cells and NK-lymphoma cells but differ at particular sites that correspond to open chromatin. oncogenic translocations lead to dysregulation of a specific set of genes controlling development. In normal T cells and in NK cells, there are hot spots of translocations at sites possessing strong H3K27ac marks.
1. Introduction
Chromosomal translocations are a physiologic mechanism of DNA recombination in germline cells and in lymphocyte development. Although these mechanisms have been studied for several decades, they are still not fully understood. Chromosomal translocations are common in cancers and more frequently occur between neighboring chromosomal regions
[1][2]. As a result, some translocations lead to the formation of oncogenic fusion proteins or to the activation of oncogenes by the altered regulatory regions
[3][4]. Ribosomal DNA (rDNA) genes are the most fragile regions in the human genome and possess nine hot spots of DSBs, each of which is about 50–100 bp in length and are denoted “Pleiades”
[5][6]. Additionally, it was demonstrated by the 4C (circular chromosome conformation capture) approach that rDNA clusters shape contacts with different chromosomal regions and are involved in the epigenetic regulation of genes that are highly associated with differentiation and cancer
[7]. Interestingly, rDNA-contacting sites often possess hot spots of DSBs. rDNA clusters also make frequent contacts with pericentromeric regions and regions possessing stretches of 5–50 kb that contain H3K27ac marks
[5]. Therefore, Pleiades may be responsible for Robertsonian translocations, in which rDNA-containing chromosomes are necessarily involved via their broken pericentromeric regions
[5]. These data indicate that rDNA genes are a candidate for mediating chromosomal translocations in normal and cancer cells. Here, we investigated whether rDNA genes are involved in chromosomal translocations by searching for rDNA-mediated translocations in normal T cells and in natural killer (NK)-cell lymphomas from the same individuals.
Natural killer cells play an important role in the innate immune system that controls several types of tumors and infections
[8]. NK cells, along with B and T lymphocytes, belong to a group of innate lymphoid cells that differentiate from a common lymphoid progenitor. NK cells are closely related to T cells and can lyse target cells without prior sensitization.
Active rDNA clusters are involved in translocations in both types of cells, particularly in regions with actively transcribed genes, but the spectra of the target genes in these cell types differ. The same rDNA-mediated hot spots of translocations are found in normal and cancer cells corresponding to the hot spots of inter-chromosomal contacts of nucleoli within specific genomic regions.
2. Chromosomal Translocations in NK-Cell Lymphomas
Translocations occur between closely located broken chromosomes. The critical distance between chromosomal regions essential for translocation is not known, but the fact that inter-chromosomal rDNA contacts, which were mapped at a resolution of ±2.5 kb
[5], are involved in translocations suggests that this space is enough for a translocation event. Many different factors are also important for translocations: the frequencies of simultaneous DNA breakage in the contacting chromosomal regions, the accessibility of DNA sequences at both sites, and the dynamics of chromosomal contacts. We observed a good correlation between translocation frequencies for all nine hot spots of DSBs in rDNA units (R1–R9) in the normal and cancer cells, although the frequencies between different hot spots vary over a wide range. The data support the view that local chromosomal states are important for translocations. The dynamics of contacts are compartment-dependent and should be different for A and B compartments that correspond to active chromatin and silent chromatin, respectively
[9]. Compartmentalization states that are formed by phase-separation mechanisms shaped by the attraction between chromatin domains of a similar state have differences in their chromatin interaction stabilities, as measured by the liquid chromatin Hi-C
[10]. Pleiades are detected only in the active rDNA units, as far as the major γ-H2AX foci and UBF-1 binding sites are co-localized in interphase nuclei
[6]. It follows that rDNA-mediated translocations should be enriched in open chromatin regions where active condensates are formed. The fact that
DUX4 genes, which are silenced and form frequent contacts with rDNA
[11][12], are not involved in rDNA-mediated translocations in either T cells or NK-cell lymphomas support this supposition.
Recently, an unexpected diversity of NK cells with distinct transcriptomes was described, including a population of NK cells associated with the expression of rDNA
[13]. The data are consistent with the regulatory role of nucleoli in the global regulation of gene expression. Previous results suggested that phase-separation mechanisms are involved in the inter-chromosomal interactions of rDNA units
[12]. Growing evidence suggests the role of rDNA units in the global regulation of genes controlling development and differentiation
[5][7][14][15][16]. We suppose that nucleoli contacts will be detected not only at the conserved multiple rDNA-contacting sites detected in different cell lines (
Figure 3) but also in the regions of detected translocations in T and NK cells. The differences in the translocation sites in these cell types may depend on the patterns of genes that shape the contacts with the nucleoli during differentiation.
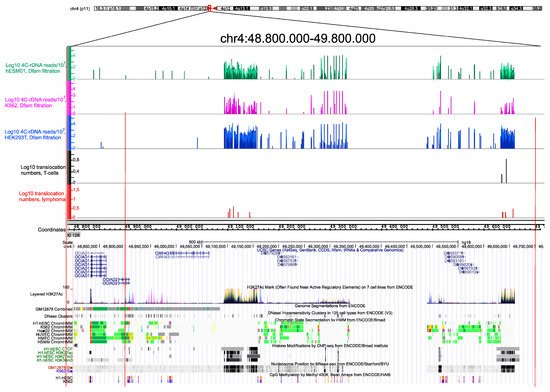
Figure 3. Translocation sites in both normal T cells and in NK-cell lymphomas coincide with and correspond to the hot spots of rDNA contacts in three human cell lines of different origin. The distribution of rDNA-contacting sites in K562 cells, HEK293T, and in hESM01 cells
[17] in the pericentromeric region of chr4 are shown as in IGB Browser (hg19). The translocation sites detected in 102 samples, each containing amplicons from T cells and NK-cell lymphomas from the same individual, are shown. The distribution of UCSC genes, layered H3K27ac marks, genome segmentation from ENCODE, histone modifications, nucleosome position, and CpG methylation inside of a 1-Mb region of chr4 are shown as in the UCSC Browser (hg19).
The etiology of NK-cell lymphoma is unknown, although there is a strong association with the Epstein–Barr virus
[18]. No specific chromosomal translocation has been identified in NK-cell lymphomas, and a multi-omics study revealed several subtypes of NK-cell lymphomas that differ in their transcriptional signatures
[19]. Three-dimensional chromosomal structures are highly variable between individual cells
[20], which suggests that heterogeneity of the chromosomal architecture in NK cells could result in differences between tumorigenic translocations and lead to several subtypes of NK-cell lymphomas with different transcriptional signatures.
Translocations inside the big intron in the
CTCF gene in NK-cell lymphomas (
Figure S1, B) might not change expression of the gene, as tested by RNA-Seq. Nevertheless, such damage of
CTCF could disrupt its function. Recently, it was shown that CTCF insulators serve as putative cancer drivers in different tumors, including lymphomas
[21].
About 100 somatic translocations per genome were detected in both T cells and NK-cell lymphomas . The majority of the genes associated with translocations did not overlap between the cell types . We conclude that in T cells, translocations were not deleterious, and a particular translocation probably only affected a subset of T cells from the donor. Of course, all translocations are subjected to selection for viability in the corresponding cells and/or in their daughter cells. As a result, only some of the translocations should be present in a cancer stem cell if the translocations, or even a single translocation, are tumorigenic. During cancer progression, rDNA-mediated translocations in daughter cells could also occur. These novel translocations may affect a specific set of sites depending on the inter-chromosomal contacts of the nucleoli in cancer cells. As result, the differences in translocation patterns between populations of T cells and NK-cancer cells should increase. We suppose that these events are responsible for the observed discrepancy between the rDNA-mediated translocations in these two cell types.
Analysis of genomic features of lung cancer patients revealed mutational signatures in B lymphocytes that are coupled with distant chromosomal rearrangements, some of which correspond to fusions involving genes with important functions
[22]. These data are consistent with our observation of translocations in normal and cancer cells. T cells and NK cells differentiate from the common lymphoid progenitor. Our data on the distinct rDNA-mediated translocation sites, which mainly originate from active chromosomal regions in T cells and NK-cell lymphomas, suggest that, even initially, the normal NK cells and T cells should have distinct patterns of expressed genes.
3. Conclusions
It was predicted that the DSB hot spots in rDNA repeats that shape frequent inter-chromosomal contact with genes controlling differentiation and development could lead to translocations that give rise to cancer transformation. Analysis of normal T cells and NK-cell lymphomas from the same individuals showed that there are rDNA-mediated translocations in both cell types. However, the translocations in normal cells and tumor cells affect different set of genes that control development.