1000/1000
Hot
Most Recent

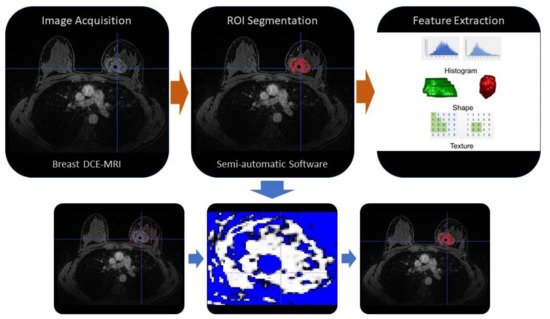
Radiomics is an emerging translational field of medicine based on the extraction of high-dimensional data from radiological images, with the purpose to reach reliable models to be applied into clinical practice for the purposes of diagnosis, prognosis and evaluation of disease response to treatment.
| Modality/Technique | Author | Purpose | Radiomics Features Category and Purpose | Population | Results | Conclusion |
|---|---|---|---|---|---|---|
| CESM | Lin et al., 2020 [35] | Identification of benign and malignant BC lesions <1 cm | Radiomics features extracted from low-energy and recombined images on CC position | 139 patients | The radiomics nomogram combined with Radiomic-score, BI-RADS category and age showed AUC of 0.940. | The radiomics nomogram incorporated with CESM-based radiomics features, BI-RADS category and age could identify benign and malignant BC <1 cm |
| CESM | Mao et al. 2020 [36] | Pre-operative prediction of ALN metastasis | LASSO logistic regression was established for feature selection and utilized to construct radiomics signature | 394 patients | ROC curves of 0.774, 0.767 and 0.79 in the training, internal validation and external validation sets, respectively. | Authors identified the cutoff score in the radiomics nomogram as −1.49, which corresponded to a total point of 49 that could diagnose ALN metastasis with a sensitivity of >95%. |
| MRI | Tan et al., 2020 [6] | Value of radiomics feature extracted on the fat-suppressed T2WI for preoperative predicting ALN metastasis in BC | 17 texture features, 5 first-order statistical features, patient age, tumor size, HER2 status and thrombus | 329 BCs | Sensitivity, specificity, accuracy and are under the curve value of radiomics signature 65.22%, 81.08%, 75.00% and 0.819. | The MRI-based radiomics signature and nomogram could be used as a non-invasive and reliable tool in predicting ALN metastasis. |
| Choudhery et al., 2020 [5] | Assessment of BC molecular subtype, pCR and Residual Cancer Burden in BC Patients Treated with NACT | Morphological and three-dimensional MRI textural features were computed, including unfiltered and filtered image data, with different spatial scaling factors | 259 BCs | Differences in minimum signal intensity and entropy among the tumor subtypes were significant. Sphericity in HER2+ tumors and entropy in luminal tumors were significantly associated with pCR. Multiple features demonstrated significant association with pathological complete response and residual cancer burden in TNBC with SD of intensity achieving the highest AUC for pCR in TNBC. | MRI radiomics features are associated with different molecular subtypes of breast cancer, pathological complete response and residual cancer burden. | |
| Hao et al., 2020 [37] | Contralateral BI-RADS 4 lesion assessment | 1046 radiomic features | 178 BCs | DCE-T1WI and T2WI imaging features signatures yielded an AUC of 0.77, which was better than the AUC of each signature alone. | The MRI radiomics-based ML model based on T2WI and DCE-T1WI features provided complementary information in discriminating benign and malignant contralateral BI-RADS 4 lesions. | |
| Lo Gullo et al., 2020 [38] | Assessment of sub-centimetric breast masses in BRCA patients | Radiomics features calculated using open-source CERR software | 96 BRCA carrier | The ML model combining 5 parameters including clinical factors, GLCM-based correlation from the pre-DCE phases and first-order coefficient of variation from the 1st post-DCE phase, achieved a diagnostic accuracy of 81.5%. | Radiomics analysis improved diagnostic accuracy compared with qualitative morphological assessment alone. | |
| Demircioglu et al., 2020 [39] | Molecular subtype, hormonal receptor status, Ki67- and HER2-expression, metastasis of lymph nodes and lymph vessel involvement as well as grading | 13.118 radiomic features extracted with a VOI-based approach | 98 BCs | PR and ER status predictions yielded AUCs of 0.67–0.69, Ki67 0.81 and HER2 Expressions 0.62. Involvement of the ALN could be predicted with an AUC of 0.80, while lymph node metastasis yielded an AUC of 0.71. | A rapid approach to VOI-based tumor-annotations for radiomics provides consisternt results to other studies in the same field. | |
| Zhang et al., 2020 [40] | Differentiation between benign and malignant lesions | Radiomics features extracted from T2WI, T1WI, DKI, ADC maps and DCE pharmacokinetic parameter maps | 207 BCs | The AUC of the optimal radiomics model, including T2 WI, DKI and quantitative DCE-MRI parameter maps was 0.921, with an accuracy of 0.833. | The model based on radiomics features from T2WI, DKI and quantitative DCE parameter maps has a high discriminatory ability for benign and malignant BC lesions. | |
| Zhou et al., 2020 [41] | Differentiation between benign and malignant BC lesions | 99 texture and histogram parameters | 133 patients | The highest accuracy of 91% was achieved when using the smallest bounding box of peritumoral tissues in segmentation. | Using the smallest bounding box containing proximal peritumor tissue as input had higher accuracy compared to using tumor alone or larger boxes. | |
| Liu et al., 2019 [42] | Assess lymphovascular invasion status | Radiomic signature composed of two features | 149 BCs | The value of AUC for a model combining both radiomic signature and ALN status (0.763) was higher than that for MRI ALN status alone and similar to that for the radiomics signature. | The DCE-MRI-based radiomics signature in combination with ALN status was effective in predicting the lymph and vascular invasion status of patients with BC before surgery. | |
| Xie et al., 2019 [43] | Subtype classification of breast cancer | 2498 features extracted from the DCE and DWI, together with DCE images, changing over 6 time points and DWI images changing over 3 b-values | 134 invasive ductal carcinoma | Highest accuracy of 91% for comparing triple negative to non-triple negative cancers. | Whole-tumor radiomics on MRI provides a non-invasive approach for BC subtype classification. | |
| Liang et al., 2018 [44] | Preoperative Ki-67 status | Radiomic features based on T2W and DCE-T1WI | 318 BC | The T2W image-based radiomics classifier showed significant discrimination for Ki-67 status, with AUC of 0.74 in the validation dataset. | The T2WI-based radiomics classifier was a significant predictor of Ki-67 status in patients with breast cancer while DCE-T1WI radiomic features were not able to discriminate Ki-67 status in the validation dataset. | |
| Digital mammography | Tan et al., 2020 [45] | Pre-operative prediction of ALN metastasis | Radiomic signature nomogram combined with receptor status and molecular subtype | 216 BCs | The radiomics nomogram, comprising PR status, molecular subtype and radiomics signature, showed excellent calibration and better performance for the metastatic ALN detection (AUC 0.883 and 0.863 in the primary and validation cohorts), better than each independent clinical feature and radiomics signature. | The mammography-based radiomics nomogram could be used as a non-invasive and reliable tool in predicting ALN metastasis. |
| Digital Mammography | Stelzer et al., 2020 [46] | Distinguish malignant from benign classification | 249 image features from gray-value histogram, co-occurrence and run-length matrices | 226 patients | A high sensitivity threshold criterion was identified in the training dataset and successfully applied to the testing dataset, demonstrating the potential to avoid 37.1-45.7 % of unnecessary biopsies at the cost of one false-negative. | Combined texture analysis and ML could be used for risk stratification in suspicious mammographic calcifications. |
| Zhou et al., 2019 [47] | HER-2 status | 186 radiomic features | 306 l BCs | In the testing set the AUC of the radiomic model in assessing HER-2 status was 0.787. | Radiomics features could help in the preoperative evaluation of HER-2 status in patients with BC. | |
| Lei et al., 2019 [48] | Prediction of benign BI-RADS 4 calcifications | 8286 radiomic features extracted from the craniocaudal and mediolateral oblique scans | 212 calcifications | Six radiomic features and the menopausal state included in a radiomic nomogram could discriminate benign from malignant calcifications with an AUC of 0.80 in the validation cohort. | The mammography-based radiomic nomogram is a potential tool to distinguish benign calcifications from malignant calcifications. | |
| PET/CT | Ou et al., 2020 [49] | Differentiating breast carcinoma from breast lymphoma | Radiomic features extracted with a local software | 44 BCs | AUCs of 0.867 and 0.806 for PET radiomic and clinical model, AUCs of 0.891 and 0.759 for CT based radiomic model on training and validation data. | Models based on clinical, and radiomic features of 18 F-FDG PET/CT images could accurately discriminate BC from breast lymphoma. |
| Reference | Modality/Techique | Purpose | Radiomics Features Category and Purpose | Population | Results | Conclusion |
|---|---|---|---|---|---|---|
| Reig et al., 2020 [50] | MRI | Review focused on machine learning techniques in breast MRI | Pre-processing, neural networks, deep learning, machine learning, segmentation, texture analysis | Breast malignant and benign pathology. | The Author discuss the possible future directions of machine learning in the current workflow of breast lesions assessed with MRI. | |
| Granzier et al., 2019 [51] | MRI | Systematic review, response prediction of neoadjuvant therapy | Various radiomic feature models, evaluated with the Radiomics Quality Score (RQS) | Studies ranging between 35-414 BC | AUC values ranged from 0.83 to 0.85. The best performing multivariate prediction model, based on logistic regression analysis, showed AUC of 0.94. | The systematic review revealed large heterogeneity for each step of the MRI-based radiomics workflow. Consequently, the results are difficult to compare. |